[English] 日本語
 Yorodumi
Yorodumi- EMDB-29940: Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | |||||||||
 Map data Map data | PGE2-EP4-Gi-map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  GPCR complex / GPCR complex /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of eosinophil extravasation /  prostaglandin E receptor activity / Prostanoid ligand receptors / negative regulation of integrin activation / response to nematode / T-helper cell differentiation / prostaglandin E receptor activity / Prostanoid ligand receptors / negative regulation of integrin activation / response to nematode / T-helper cell differentiation /  regulation of stress fiber assembly / negative regulation of cytokine production / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex ...negative regulation of eosinophil extravasation / regulation of stress fiber assembly / negative regulation of cytokine production / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex ...negative regulation of eosinophil extravasation /  prostaglandin E receptor activity / Prostanoid ligand receptors / negative regulation of integrin activation / response to nematode / T-helper cell differentiation / prostaglandin E receptor activity / Prostanoid ligand receptors / negative regulation of integrin activation / response to nematode / T-helper cell differentiation /  regulation of stress fiber assembly / negative regulation of cytokine production / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex / regulation of stress fiber assembly / negative regulation of cytokine production / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex /  regulation of ossification / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / response to mechanical stimulus / regulation of mitotic spindle organization / cellular response to forskolin / JNK cascade / ERK1 and ERK2 cascade / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Regulation of insulin secretion / G protein-coupled receptor binding / positive regulation of cytokine production / Olfactory Signaling Pathway / G-protein beta/gamma-subunit complex binding / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / regulation of ossification / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / response to mechanical stimulus / regulation of mitotic spindle organization / cellular response to forskolin / JNK cascade / ERK1 and ERK2 cascade / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Regulation of insulin secretion / G protein-coupled receptor binding / positive regulation of cytokine production / Olfactory Signaling Pathway / G-protein beta/gamma-subunit complex binding / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / adenylate cyclase-modulating G protein-coupled receptor signaling pathway /  bone development / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / adenylate cyclase-activating G protein-coupled receptor signaling pathway / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / response to peptide hormone / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / negative regulation of inflammatory response / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / positive regulation of inflammatory response / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to mechanical stimulus / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / GDP binding / bone development / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / adenylate cyclase-activating G protein-coupled receptor signaling pathway / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Sensory perception of sweet, bitter, and umami (glutamate) taste / response to peptide hormone / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / negative regulation of inflammatory response / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / positive regulation of inflammatory response / ADORA2B mediated anti-inflammatory cytokines production / adenylate cyclase-activating dopamine receptor signaling pathway / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / cellular response to mechanical stimulus / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / sensory perception of taste / GPER1 signaling / G-protein beta-subunit binding / GDP binding /  heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events /  extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye /  GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / GTPase binding / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway /  cell cortex / midbody / positive regulation of cytosolic calcium ion concentration / G alpha (i) signalling events / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / response to lipopolysaccharide / Extra-nuclear estrogen signaling / cell cortex / midbody / positive regulation of cytosolic calcium ion concentration / G alpha (i) signalling events / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / response to lipopolysaccharide / Extra-nuclear estrogen signaling /  immune response / immune response /  inflammatory response / inflammatory response /  cell cycle / G protein-coupled receptor signaling pathway / lysosomal membrane / cell cycle / G protein-coupled receptor signaling pathway / lysosomal membrane /  cell division / cell division /  GTPase activity / GTPase activity /  centrosome / centrosome /  synapse / protein-containing complex binding / GTP binding / synapse / protein-containing complex binding / GTP binding /  nucleolus / magnesium ion binding / nucleolus / magnesium ion binding /  signal transduction / extracellular exosome / signal transduction / extracellular exosome /  nucleoplasm nucleoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.1 Å cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Huang SM / Xiong MY / Liu L / Mu J / Sheng C / Sun J | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Single hormone or synthetic agonist induces G/G coupling selectivity of EP receptors via distinct binding modes and propagating paths. Authors: Shen-Ming Huang / Meng-Yao Xiong / Lei Liu / Jianqiang Mu / Ming-Wei Wang / Ying-Li Jia / Kui Cai / Lu Tie / Chao Zhang / Sheng Cao / Xin Wen / Jia-Le Wang / Sheng-Chao Guo / Yu Li / Chang- ...Authors: Shen-Ming Huang / Meng-Yao Xiong / Lei Liu / Jianqiang Mu / Ming-Wei Wang / Ying-Li Jia / Kui Cai / Lu Tie / Chao Zhang / Sheng Cao / Xin Wen / Jia-Le Wang / Sheng-Chao Guo / Yu Li / Chang-Xiu Qu / Qing-Tao He / Bo-Yang Cai / Chenyang Xue / Shiyi Gan / Yihe Xie / Xin Cong / Zhao Yang / Wei Kong / Shuo Li / Zijian Li / Peng Xiao / Fan Yang / Xiao Yu / You-Fei Guan / Xiaoyan Zhang / Zhongmin Liu / Bao-Xue Yang / Yang Du / Jin-Peng Sun /  Abstract: To accomplish concerted physiological reactions, nature has diversified functions of a single hormone at at least two primary levels: 1) Different receptors recognize the same hormone, and 2) ...To accomplish concerted physiological reactions, nature has diversified functions of a single hormone at at least two primary levels: 1) Different receptors recognize the same hormone, and 2) different cellular effectors couple to the same hormone-receptor pair [R.P. Xiao, , re15 (2001); L. Hein, J. D. Altman, B.K. Kobilka, , 181-184 (1999); Y. Daaka, L. M. Luttrell, R. J. Lefkowitz, , 88-91 (1997)]. Not only these questions lie in the heart of hormone actions and receptor signaling but also dissecting mechanisms underlying these questions could offer therapeutic routes for refractory diseases, such as kidney injury (KI) or X-linked nephrogenic diabetes insipidus (NDI). Here, we identified that G-biased signaling, but not G activation downstream of EP4, showed beneficial effects for both KI and NDI treatments. Notably, by solving Cryo-electron microscope (cryo-EM) structures of EP3-G, EP4-G, and EP4-G in complex with endogenous prostaglandin E (PGE)or two synthetic agonists and comparing with PGE-EP2-G structures, we found that unique primary sequences of prostaglandin E2 receptor (EP) receptors and distinct conformational states of the EP4 ligand pocket govern the G/G transducer coupling selectivity through different structural propagation paths, especially via TM6 and TM7, to generate selective cytoplasmic structural features. In particular, the orientation of the PGE ω-chain and two distinct pockets encompassing agonist L902688 of EP4 were differentiated by their G/G coupling ability. Further, we identified common and distinct features of cytoplasmic side of EP receptors for G/G coupling and provide a structural basis for selective and biased agonist design of EP4 with therapeutic potential. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29940.map.gz emd_29940.map.gz | 32.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29940-v30.xml emd-29940-v30.xml emd-29940.xml emd-29940.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29940.png emd_29940.png | 89.6 KB | ||
| Masks |  emd_29940_msk_1.map emd_29940_msk_1.map | 38.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29940.cif.gz emd-29940.cif.gz | 7.1 KB | ||
| Others |  emd_29940_half_map_1.map.gz emd_29940_half_map_1.map.gz emd_29940_half_map_2.map.gz emd_29940_half_map_2.map.gz | 29.6 MB 29.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29940 http://ftp.pdbj.org/pub/emdb/structures/EMD-29940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29940 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29940 | HTTPS FTP |
-Related structure data
| Related structure data |  8gcpMC  8gcmC  8gd9C  8gdaC  8gdbC  8gdcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29940.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29940.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PGE2-EP4-Gi-map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29940_msk_1.map emd_29940_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: PGE2-EP4-Gi-map-half1
| File | emd_29940_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PGE2-EP4-Gi-map-half1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: PGE2-EP4-Gi-map-half2
| File | emd_29940_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PGE2-EP4-Gi-map-half2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM Structure of Prostaglandin E Receptor EP4 Coupled to G Protein
| Entire | Name: Cryo-EM Structure of Prostaglandin E Receptor EP4 Coupled to G Protein |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM Structure of Prostaglandin E Receptor EP4 Coupled to G Protein
| Supramolecule | Name: Cryo-EM Structure of Prostaglandin E Receptor EP4 Coupled to G Protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.445059 KDa |
| Recombinant expression | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein subunit gamma
| Macromolecule | Name: Guanine nucleotide-binding protein subunit gamma / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.104898 KDa |
| Recombinant expression | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MWRELPLGLG ELHKDHQASR KLEPELWSVS ENPPSTSMAS NNTASIAQAR KLVEQLKMEA NIDRIKVSKA AADLMAYCEA HAKEDPLLT PVPASENPFR EKKFFCAIL UniProtKB: Guanine nucleotide-binding protein subunit gamma |
-Macromolecule #4: scFv
| Macromolecule | Name: scFv / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.898781 KDa |
| Recombinant expression | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFADV QLVESGGGLV QPGGSRKLSC SASGFAFSSF GMHWVRQAPE KGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSEDT AMYYCVRSIY YYGSSPFDFW GQGTTLTVSS G GGGSGGGG ...String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFADV QLVESGGGLV QPGGSRKLSC SASGFAFSSF GMHWVRQAPE KGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSEDT AMYYCVRSIY YYGSSPFDFW GQGTTLTVSS G GGGSGGGG SGGGGSDIVM TQATSSVPVT PGESVSISCR SSKSLLHSNG NTYLYWFLQR PGQSPQLLIY RMSNLASGVP DR FSGSGSG TAFTLTISRL EAEDVGVYYC MQHLEYPLTF GAGTKLELKG SLEVLFQGPA AAHHHHHHHH |
-Macromolecule #5: Prostaglandin E2 receptor EP4 subtype
| Macromolecule | Name: Prostaglandin E2 receptor EP4 subtype / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 53.173336 KDa |
| Recombinant expression | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: MSTPGVNSSA SLSPDRLNSP VTIPAVMFIF GVVGNLVAIV VLCKSRKEQK ETTFYTLVCG LAVTDLLGTL LVSPVTIATY MKGQWPGGQ PLCEYSTFIL LFFSLSGLSI ICAMSVERYL AINHAYFYSH YVDKRLAGLT LFAVYASNVL FCALPNMGLG S SRLQYPDT ...String: MSTPGVNSSA SLSPDRLNSP VTIPAVMFIF GVVGNLVAIV VLCKSRKEQK ETTFYTLVCG LAVTDLLGTL LVSPVTIATY MKGQWPGGQ PLCEYSTFIL LFFSLSGLSI ICAMSVERYL AINHAYFYSH YVDKRLAGLT LFAVYASNVL FCALPNMGLG S SRLQYPDT WCFIDWTTNV TAHAAYSYMY AGFSSFLILA TVLCNVLVCG ALLRMHRQFM RRTSLGTEQH HAAAAASVAS RG HPAASPA LPRLSDFRRR RSFRRIAGAE IQMVILLIAT SLVVLICSIP LVVRVFVNQL YQPSLEREVS KNPDLQAIRI ASV NPILDP WIYILLRKTV LSKAIEKIKC LFCRIGGSRR ERSGQHCSDS QRTSSAMSGH SRSFISRELK EISSTSQTLL PDLS LPDLS ENGLGGRNLL PGVPGMGLAQ EDTTSLRTLR ISETSDSSQG QDSESVLLVD EAGGSGRAGP APKGSSLQVT FPSET LNLS EKCI UniProtKB: Prostaglandin E2 receptor EP4 subtype |
-Macromolecule #6: (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-...
| Macromolecule | Name: (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid type: ligand / ID: 6 / Number of copies: 1 / Formula: P2E |
|---|---|
| Molecular weight | Theoretical: 352.465 Da |
| Chemical component information | 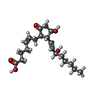 ChemComp-P2E: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 25.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: PROJECTION MATCHING |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 291813 |
 Movie
Movie Controller
Controller




































 Z
Z Y
Y X
X


























