[English] 日本語
 Yorodumi
Yorodumi- PDB-8g5h: Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8g5h | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroid | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  ligand-gated ion channel / ligand-gated ion channel /  cys-loop receptor / cys-loop receptor /  neurotransmitter receptor neurotransmitter receptor | |||||||||
| Function / homology |  Function and homology information Function and homology informationGABA receptor activation /  diazepam binding / diazepam binding /  inhibitory synapse / inhibitory synapse /  GABA receptor complex / GABA receptor complex /  organic cyclic compound binding / inhibitory extracellular ligand-gated monoatomic ion channel activity / inner ear receptor cell development / GABA-gated chloride ion channel activity / cellular response to histamine / organic cyclic compound binding / inhibitory extracellular ligand-gated monoatomic ion channel activity / inner ear receptor cell development / GABA-gated chloride ion channel activity / cellular response to histamine /  inhibitory synapse assembly ...GABA receptor activation / inhibitory synapse assembly ...GABA receptor activation /  diazepam binding / diazepam binding /  inhibitory synapse / inhibitory synapse /  GABA receptor complex / GABA receptor complex /  organic cyclic compound binding / inhibitory extracellular ligand-gated monoatomic ion channel activity / inner ear receptor cell development / GABA-gated chloride ion channel activity / cellular response to histamine / organic cyclic compound binding / inhibitory extracellular ligand-gated monoatomic ion channel activity / inner ear receptor cell development / GABA-gated chloride ion channel activity / cellular response to histamine /  inhibitory synapse assembly / inhibitory synapse assembly /  GABA-A receptor activity / GABA-A receptor activity /  GABA-A receptor complex / GABA-A receptor complex /  innervation / innervation /  neurotransmitter receptor activity / postsynaptic specialization membrane / gamma-aminobutyric acid signaling pathway / neurotransmitter receptor activity / postsynaptic specialization membrane / gamma-aminobutyric acid signaling pathway /  synaptic transmission, GABAergic / synaptic transmission, GABAergic /  adult behavior / adult behavior /  chloride channel activity / cochlea development / chloride channel activity / cochlea development /  chloride channel complex / GABA-ergic synapse / neuron development / regulation of postsynaptic membrane potential / presynaptic active zone membrane / chloride transmembrane transport / dendrite membrane / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / post-embryonic development / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / cytoplasmic vesicle membrane / chemical synaptic transmission / cytoplasmic vesicle / postsynapse / neuron projection / chloride channel complex / GABA-ergic synapse / neuron development / regulation of postsynaptic membrane potential / presynaptic active zone membrane / chloride transmembrane transport / dendrite membrane / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / post-embryonic development / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / cytoplasmic vesicle membrane / chemical synaptic transmission / cytoplasmic vesicle / postsynapse / neuron projection /  axon / glutamatergic synapse / axon / glutamatergic synapse /  synapse / synapse /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.89 Å cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Sun, C. / Gouaux, E. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Cryo-EM structures reveal native GABA receptor assemblies and pharmacology. Authors: Chang Sun / Hongtao Zhu / Sarah Clark / Eric Gouaux /   Abstract: Type A γ-aminobutyric acid receptors (GABARs) are the principal inhibitory receptors in the brain and the target of a wide range of clinical agents, including anaesthetics, sedatives, hypnotics and ...Type A γ-aminobutyric acid receptors (GABARs) are the principal inhibitory receptors in the brain and the target of a wide range of clinical agents, including anaesthetics, sedatives, hypnotics and antidepressants. However, our understanding of GABAR pharmacology has been hindered by the vast number of pentameric assemblies that can be derived from 19 different subunits and the lack of structural knowledge of clinically relevant receptors. Here, we isolate native murine GABAR assemblies containing the widely expressed α1 subunit and elucidate their structures in complex with drugs used to treat insomnia (zolpidem (ZOL) and flurazepam) and postpartum depression (the neurosteroid allopregnanolone (APG)). Using cryo-electron microscopy (cryo-EM) analysis and single-molecule photobleaching experiments, we uncover three major structural populations in the brain: the canonical α1β2γ2 receptor containing two α1 subunits, and two assemblies containing one α1 and either an α2 or α3 subunit, in which the single α1-containing receptors feature a more compact arrangement between the transmembrane and extracellular domains. Interestingly, APG is bound at the transmembrane α/β subunit interface, even when not added to the sample, revealing an important role for endogenous neurosteroids in modulating native GABARs. Together with structurally engaged lipids, neurosteroids produce global conformational changes throughout the receptor that modify the ion channel pore and the binding sites for GABA and insomnia medications. Our data reveal the major α1-containing GABAR assemblies, bound with endogenous neurosteroid, thus defining a structural landscape from which subtype-specific drugs can be developed. #1: Journal: Acta Crystallogr D Struct Biol / Year: 2018 Title: Real-space refinement in PHENIX for cryo-EM and crystallography. Authors: Pavel V Afonine / Billy K Poon / Randy J Read / Oleg V Sobolev / Thomas C Terwilliger / Alexandre Urzhumtsev / Paul D Adams /    Abstract: This article describes the implementation of real-space refinement in the phenix.real_space_refine program from the PHENIX suite. The use of a simplified refinement target function enables very fast ...This article describes the implementation of real-space refinement in the phenix.real_space_refine program from the PHENIX suite. The use of a simplified refinement target function enables very fast calculation, which in turn makes it possible to identify optimal data-restraint weights as part of routine refinements with little runtime cost. Refinement of atomic models against low-resolution data benefits from the inclusion of as much additional information as is available. In addition to standard restraints on covalent geometry, phenix.real_space_refine makes use of extra information such as secondary-structure and rotamer-specific restraints, as well as restraints or constraints on internal molecular symmetry. The re-refinement of 385 cryo-EM-derived models available in the Protein Data Bank at resolutions of 6 Å or better shows significant improvement of the models and of the fit of these models to the target maps. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8g5h.cif.gz 8g5h.cif.gz | 428.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8g5h.ent.gz pdb8g5h.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  8g5h.json.gz 8g5h.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g5/8g5h https://data.pdbj.org/pub/pdb/validation_reports/g5/8g5h ftp://data.pdbj.org/pub/pdb/validation_reports/g5/8g5h ftp://data.pdbj.org/pub/pdb/validation_reports/g5/8g5h | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  29743MC  8foiC  8g4nC  8g4oC  8g4xC  8g5fC  8g5gC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Gamma-aminobutyric acid receptor subunit ... , 4 types, 5 molecules BECDA
| #1: Protein | Mass: 59265.312 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   Mus musculus (house mouse) / Strain: C57BL/6 / References: UniProt: P63137 Mus musculus (house mouse) / Strain: C57BL/6 / References: UniProt: P63137#2: Protein | | Mass: 51818.516 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Mus musculus (house mouse) / Strain: C57BL/6 / References: UniProt: P62812 Mus musculus (house mouse) / Strain: C57BL/6 / References: UniProt: P62812#3: Protein | | Mass: 55160.281 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Mus musculus (house mouse) / Strain: C57BL/6 / References: UniProt: P22723 Mus musculus (house mouse) / Strain: C57BL/6 / References: UniProt: P22723#6: Protein | | Mass: 55461.684 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Mus musculus (house mouse) / Strain: C57BL/6 / References: UniProt: P26049 Mus musculus (house mouse) / Strain: C57BL/6 / References: UniProt: P26049 |
|---|
-Antibody , 2 types, 2 molecules JK
| #4: Antibody | Mass: 24159.895 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Production host: Mus musculus (house mouse) / Production host:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
|---|---|
| #5: Antibody | Mass: 23624.080 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Production host: Mus musculus (house mouse) / Production host:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm) |
-Sugars , 4 types, 8 molecules 
| #7: Polysaccharide | alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2- ...alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose  / Mass: 910.823 Da / Num. of mol.: 5 / Mass: 910.823 Da / Num. of mol.: 5Source method: isolated from a genetically manipulated source #8: Polysaccharide | alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D- ...alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | Type: oligosaccharide  / Mass: 1219.105 Da / Num. of mol.: 1 / Mass: 1219.105 Da / Num. of mol.: 1Source method: isolated from a genetically manipulated source #9: Polysaccharide | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta- ...beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose |  / Mass: 586.542 Da / Num. of mol.: 1 / Source method: obtained synthetically / Mass: 586.542 Da / Num. of mol.: 1 / Source method: obtained synthetically#13: Sugar | ChemComp-NAG / |  N-Acetylglucosamine N-Acetylglucosamine |
|---|
-Non-polymers , 4 types, 6 molecules 

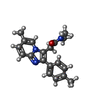
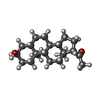



| #10: Chemical |  Γ-Aminobutyric acid Γ-Aminobutyric acid#11: Chemical | #12: Chemical | ChemComp-R5R / |  Zolpidem Zolpidem#14: Chemical | ChemComp-Y4B / | |
|---|
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Native GABAA Receptor isolated from mouse brain containing one alpha1 subunit, in complex with 8E3-Fab, GABA, Zolpidem, and endogenous neurosteroid Type: COMPLEX / Entity ID: #1-#6 / Source: NATURAL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.35 MDa / Experimental value: NO | |||||||||||||||
| Source (natural) | Organism:   Mus musculus (house mouse) / Strain: C57BL/6 / Organ: Brain Mus musculus (house mouse) / Strain: C57BL/6 / Organ: Brain | |||||||||||||||
| Buffer solution | pH: 8 | |||||||||||||||
| Buffer component |
| |||||||||||||||
| Specimen | Conc.: 0.1 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES | |||||||||||||||
| Specimen support | Grid material: GOLD / Grid mesh size: 200 divisions/in. / Grid type: Quantifoil R2/1 | |||||||||||||||
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 293 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2100 nm / Nominal defocus min: 800 nm Bright-field microscopy / Nominal defocus max: 2100 nm / Nominal defocus min: 800 nm |
| Image recording | Electron dose: 50 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| Software | Name: PHENIX / Version: 1.20.1_4487: / Classification: refinement |
|---|---|
| EM software | Name: cryoSPARC |
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
| Symmetry | Point symmetry : C1 (asymmetric) : C1 (asymmetric) |
3D reconstruction | Resolution: 2.89 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 91747 / Algorithm: BACK PROJECTION / Symmetry type: POINT |
| Atomic model building | Protocol: AB INITIO MODEL / Space: REAL |
| Refinement | Highest resolution: 2.89 Å |
 Movie
Movie Controller
Controller








 PDBj
PDBj








