[English] 日本語
 Yorodumi
Yorodumi- PDB-8c29: Cryo-EM structure of photosystem II C2S2 supercomplex from Norway... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8c29 | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of photosystem II C2S2 supercomplex from Norway spruce (Picea abies) at 2.8 Angstrom resolution | ||||||||||||||||||||||||||||||
 Components Components |
| ||||||||||||||||||||||||||||||
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  Membrane protein complex / Membrane protein complex /  Photosynthesis / Photosynthesis /  Photosystem / Photosystem /  Gymnosperm Gymnosperm | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information photosynthesis, light harvesting / photosynthesis, light harvesting /  photosystem II assembly / photosystem II assembly /  photosystem II stabilization / oxygen evolving activity / photosystem II stabilization / oxygen evolving activity /  photosystem II oxygen evolving complex / photosystem II oxygen evolving complex /  photosystem II / photosystem II /  photosystem II reaction center / photosynthetic electron transport chain / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / photosystem II reaction center / photosynthetic electron transport chain / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor /  photosystem I ... photosystem I ... photosynthesis, light harvesting / photosynthesis, light harvesting /  photosystem II assembly / photosystem II assembly /  photosystem II stabilization / oxygen evolving activity / photosystem II stabilization / oxygen evolving activity /  photosystem II oxygen evolving complex / photosystem II oxygen evolving complex /  photosystem II / photosystem II /  photosystem II reaction center / photosynthetic electron transport chain / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor / photosystem II reaction center / photosynthetic electron transport chain / oxidoreductase activity, acting on diphenols and related substances as donors, oxygen as acceptor /  photosystem I / response to herbicide / photosystem I / response to herbicide /  photosystem II / photosynthetic electron transport in photosystem II / photosystem II / photosynthetic electron transport in photosystem II /  chlorophyll binding / chloroplast thylakoid membrane / chlorophyll binding / chloroplast thylakoid membrane /  photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity / photosynthesis, light reaction / electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis activity /  phosphate ion binding / phosphate ion binding /  photosynthesis / photosynthesis /  electron transfer activity / protein stabilization / iron ion binding / electron transfer activity / protein stabilization / iron ion binding /  heme binding / heme binding /  metal ion binding metal ion bindingSimilarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |   Picea abies (Norway spruce) Picea abies (Norway spruce) | ||||||||||||||||||||||||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.785 Å cryo EM / Resolution: 2.785 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Kopecny, D. / Semchonok, D.A. / Kouril, R. | ||||||||||||||||||||||||||||||
| Funding support |  Czech Republic, Czech Republic,  Germany, 9items Germany, 9items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2023 Journal: Nat Plants / Year: 2023Title: Cryo-EM structure of a plant photosystem II supercomplex with light-harvesting protein Lhcb8 and α-tocopherol. Authors: Monika Opatíková / Dmitry A Semchonok / David Kopečný / Petr Ilík / Pavel Pospíšil / Iva Ilíková / Pavel Roudnický / Sanja Ćavar Zeljković / Petr Tarkowski / Fotis L Kyrilis / ...Authors: Monika Opatíková / Dmitry A Semchonok / David Kopečný / Petr Ilík / Pavel Pospíšil / Iva Ilíková / Pavel Roudnický / Sanja Ćavar Zeljković / Petr Tarkowski / Fotis L Kyrilis / Farzad Hamdi / Panagiotis L Kastritis / Roman Kouřil /    Abstract: The heart of oxygenic photosynthesis is the water-splitting photosystem II (PSII), which forms supercomplexes with a variable amount of peripheral trimeric light-harvesting complexes (LHCII). Our ...The heart of oxygenic photosynthesis is the water-splitting photosystem II (PSII), which forms supercomplexes with a variable amount of peripheral trimeric light-harvesting complexes (LHCII). Our knowledge of the structure of green plant PSII supercomplex is based on findings obtained from several representatives of green algae and flowering plants; however, data from a non-flowering plant are currently missing. Here we report a cryo-electron microscopy structure of PSII supercomplex from spruce, a representative of non-flowering land plants, at 2.8 Å resolution. Compared with flowering plants, PSII supercomplex in spruce contains an additional Ycf12 subunit, Lhcb4 protein is replaced by Lhcb8, and trimeric LHCII is present as a homotrimer of Lhcb1. Unexpectedly, we have found α-tocopherol (α-Toc)/α-tocopherolquinone (α-TQ) at the boundary between the LHCII trimer and the inner antenna CP43. The molecule of α-Toc/α-TQ is located close to chlorophyll a614 of one of the Lhcb1 proteins and its chromanol/quinone head is exposed to the thylakoid lumen. The position of α-Toc in PSII supercomplex makes it an ideal candidate for the sensor of excessive light, as α-Toc can be oxidized to α-TQ by high-light-induced singlet oxygen at low lumenal pH. The molecule of α-TQ appears to shift slightly into the PSII supercomplex, which could trigger important structure-functional modifications in PSII supercomplex. Inspection of the previously reported cryo-electron microscopy maps of PSII supercomplexes indicates that α-Toc/α-TQ can be present at the same site also in PSII supercomplexes from flowering plants, but its identification in the previous studies has been hindered by insufficient resolution. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8c29.cif.gz 8c29.cif.gz | 1.7 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8c29.ent.gz pdb8c29.ent.gz | 1.5 MB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8c29.json.gz 8c29.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c2/8c29 https://data.pdbj.org/pub/pdb/validation_reports/c2/8c29 ftp://data.pdbj.org/pub/pdb/validation_reports/c2/8c29 ftp://data.pdbj.org/pub/pdb/validation_reports/c2/8c29 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  16389MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Photosystem II ... , 14 types, 28 molecules AaBbCcDdHhIiKkLlMmTtUuVvXxZz
| #1: Protein |  / PSII D1 protein / Photosystem II Q(B) protein / PSII D1 protein / Photosystem II Q(B) proteinMass: 38803.215 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbA / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: P50155, Picea abies (Norway spruce) / References: UniProt: P50155,  photosystem II photosystem II#2: Protein |  Mass: 56375.496 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbB / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGX1 Picea abies (Norway spruce) / References: UniProt: R4ZGX1#3: Protein |  Mass: 51785.270 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbC / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGZ0 Picea abies (Norway spruce) / References: UniProt: R4ZGZ0#4: Protein |  / PSII D2 protein / Photosystem Q(A) protein / PSII D2 protein / Photosystem Q(A) proteinMass: 39483.117 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbD / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGX6, Picea abies (Norway spruce) / References: UniProt: R4ZGX6,  photosystem II photosystem II#8: Protein |  / PSII-H / PSII-HMass: 7941.112 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbH / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGS7 Picea abies (Norway spruce) / References: UniProt: R4ZGS7#9: Protein/peptide |  / PSII-I / PSII 4.8 kDa protein / PSII-I / PSII 4.8 kDa proteinMass: 4184.984 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbI / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGS4 Picea abies (Norway spruce) / References: UniProt: R4ZGS4#10: Protein |  Mass: 6804.083 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbK / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGX8 Picea abies (Norway spruce) / References: UniProt: R4ZGX8#11: Protein/peptide |  / PSII-L / PSII-LMass: 4475.153 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbL / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGV2 Picea abies (Norway spruce) / References: UniProt: R4ZGV2#12: Protein/peptide |  Mass: 3952.636 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbM / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: A2CHR7 Picea abies (Norway spruce) / References: UniProt: A2CHR7#16: Protein/peptide |  / PSII-T / PSII-TMass: 3962.781 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbTc / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGU2 Picea abies (Norway spruce) / References: UniProt: R4ZGU2#17: Protein |  Mass: 14189.462 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbTn / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: A9NJW3 Picea abies (Norway spruce) / References: UniProt: A9NJW3#18: Protein/peptide |  Mass: 3490.290 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: Psb30 / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGY5 Picea abies (Norway spruce) / References: UniProt: R4ZGY5#20: Protein |  Mass: 12956.940 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbX / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: A9NTS2 Picea abies (Norway spruce) / References: UniProt: A9NTS2#21: Protein |  Mass: 6412.558 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbZ / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGT1 Picea abies (Norway spruce) / References: UniProt: R4ZGT1 |
|---|
-Cytochrome b559 subunit ... , 2 types, 4 molecules EeFf
| #5: Protein |  / PSII reaction center subunit V / PSII reaction center subunit VMass: 9478.562 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbE / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGW6 Picea abies (Norway spruce) / References: UniProt: R4ZGW6#6: Protein/peptide |  / PSII reaction center subunit VI / PSII reaction center subunit VIMass: 4476.271 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbF / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: R4ZGT7 Picea abies (Norway spruce) / References: UniProt: R4ZGT7 |
|---|
-Chlorophyll a-b binding protein, ... , 3 types, 10 molecules GNYgnyRrSs
| #7: Protein | Mass: 29112.004 Da / Num. of mol.: 6 / Source method: isolated from a natural source / Details: Lhcb1 / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: Q40771 Picea abies (Norway spruce) / References: UniProt: Q40771#14: Protein | Mass: 32767.854 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: Lhcb4.3 / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: A9NKX0 Picea abies (Norway spruce) / References: UniProt: A9NKX0#15: Protein | Mass: 32287.926 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: CP26 / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: A9NKM0 Picea abies (Norway spruce) / References: UniProt: A9NKM0 |
|---|
-Protein , 2 types, 4 molecules OoWw
| #13: Protein | Mass: 36089.457 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbO / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: A9NRV4 Picea abies (Norway spruce) / References: UniProt: A9NRV4#19: Protein | Mass: 15266.507 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Details: PsbW / Source: (natural)   Picea abies (Norway spruce) / References: UniProt: A9NLC2 Picea abies (Norway spruce) / References: UniProt: A9NLC2 |
|---|
-Sugars , 1 types, 4 molecules 
| #33: Sugar | ChemComp-DGD / |
|---|
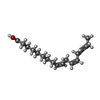
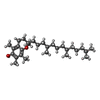
+Non-polymers , 21 types, 634 molecules 






































 Movie
Movie Controller
Controller


 PDBj
PDBj




























