[English] 日本語
 Yorodumi
Yorodumi- PDB-7z78: Crystal structure of compound 4 in complex with the bromodomain o... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7z78 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of compound 4 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | ||||||
 Components Components | Probable global transcription activator SNF2L2 | ||||||
 Keywords Keywords |  LIGASE / LIGASE /  PROTAC / PROTAC /  Complex / Complex /  Bromodomain Bromodomain | ||||||
| Function / homology |  Function and homology information Function and homology informationbBAF complex / npBAF complex /  brahma complex / brahma complex /  nBAF complex / GBAF complex / regulation of G0 to G1 transition / regulation of nucleotide-excision repair / nBAF complex / GBAF complex / regulation of G0 to G1 transition / regulation of nucleotide-excision repair /  SWI/SNF complex / ATP-dependent chromatin remodeler activity / regulation of mitotic metaphase/anaphase transition ...bBAF complex / npBAF complex / SWI/SNF complex / ATP-dependent chromatin remodeler activity / regulation of mitotic metaphase/anaphase transition ...bBAF complex / npBAF complex /  brahma complex / brahma complex /  nBAF complex / GBAF complex / regulation of G0 to G1 transition / regulation of nucleotide-excision repair / nBAF complex / GBAF complex / regulation of G0 to G1 transition / regulation of nucleotide-excision repair /  SWI/SNF complex / ATP-dependent chromatin remodeler activity / regulation of mitotic metaphase/anaphase transition / positive regulation of double-strand break repair / positive regulation of T cell differentiation / intermediate filament cytoskeleton / positive regulation of stem cell population maintenance / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / spermatid development / regulation of G1/S transition of mitotic cell cycle / negative regulation of cell differentiation / positive regulation of myoblast differentiation / ATP-dependent activity, acting on DNA / SWI/SNF complex / ATP-dependent chromatin remodeler activity / regulation of mitotic metaphase/anaphase transition / positive regulation of double-strand break repair / positive regulation of T cell differentiation / intermediate filament cytoskeleton / positive regulation of stem cell population maintenance / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / spermatid development / regulation of G1/S transition of mitotic cell cycle / negative regulation of cell differentiation / positive regulation of myoblast differentiation / ATP-dependent activity, acting on DNA /  helicase activity / positive regulation of cell differentiation / helicase activity / positive regulation of cell differentiation /  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / negative regulation of cell growth / RMTs methylate histone arginines / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / negative regulation of cell growth / RMTs methylate histone arginines /  nervous system development / nervous system development /  histone binding / histone binding /  transcription coactivator activity / transcription cis-regulatory region binding / transcription coactivator activity / transcription cis-regulatory region binding /  hydrolase activity / hydrolase activity /  chromatin remodeling / negative regulation of cell population proliferation / intracellular membrane-bounded organelle / negative regulation of DNA-templated transcription / chromatin remodeling / negative regulation of cell population proliferation / intracellular membrane-bounded organelle / negative regulation of DNA-templated transcription /  chromatin binding / positive regulation of cell population proliferation / chromatin binding / positive regulation of cell population proliferation /  chromatin / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II / chromatin / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / positive regulation of transcription by RNA polymerase II /  DNA binding / DNA binding /  nucleoplasm / nucleoplasm /  ATP binding / ATP binding /  nucleus nucleusSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.32 Å MOLECULAR REPLACEMENT / Resolution: 1.32 Å | ||||||
 Authors Authors | Bader, G. / Boettcher, J. / Wolkerstorfer, B. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo. Authors: Kofink, C. / Trainor, N. / Mair, B. / Wohrle, S. / Wurm, M. / Mischerikow, N. / Roy, M.J. / Bader, G. / Greb, P. / Garavel, G. / Diers, E. / McLennan, R. / Whitworth, C. / Vetma, V. / ...Authors: Kofink, C. / Trainor, N. / Mair, B. / Wohrle, S. / Wurm, M. / Mischerikow, N. / Roy, M.J. / Bader, G. / Greb, P. / Garavel, G. / Diers, E. / McLennan, R. / Whitworth, C. / Vetma, V. / Rumpel, K. / Scharnweber, M. / Fuchs, J.E. / Gerstberger, T. / Cui, Y. / Gremel, G. / Chetta, P. / Hopf, S. / Budano, N. / Rinnenthal, J. / Gmaschitz, G. / Mayer, M. / Koegl, M. / Ciulli, A. / Weinstabl, H. / Farnaby, W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7z78.cif.gz 7z78.cif.gz | 163.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7z78.ent.gz pdb7z78.ent.gz | 128.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7z78.json.gz 7z78.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/z7/7z78 https://data.pdbj.org/pub/pdb/validation_reports/z7/7z78 ftp://data.pdbj.org/pub/pdb/validation_reports/z7/7z78 ftp://data.pdbj.org/pub/pdb/validation_reports/z7/7z78 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7z6lC  7z76C  7z77C 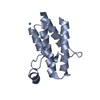 4qy4S  5t35S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 14380.542 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: SMARCA2, BAF190B, BRM, SNF2A, SNF2L2 / Production host: Homo sapiens (human) / Gene: SMARCA2, BAF190B, BRM, SNF2A, SNF2L2 / Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli)References: UniProt: P51531,  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement#2: Chemical | #3: Chemical | ChemComp-ZN / #4: Water | ChemComp-HOH / |  Water WaterHas ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.23 Å3/Da / Density % sol: 44.78 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop Details: 8% ethylene glycol, 25% PEG 6000, 0.1 M HEPES pH 8.0 and 10 mM zinc chloride |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X10SA / Wavelength: 0.9999 Å / Beamline: X10SA / Wavelength: 0.9999 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Sep 7, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9999 Å / Relative weight: 1 : 0.9999 Å / Relative weight: 1 |
| Reflection | Resolution: 1.32→89.27 Å / Num. obs: 60981 / % possible obs: 94 % / Redundancy: 5.1 % / CC1/2: 0.999 / Net I/σ(I): 16.3 |
| Reflection shell | Resolution: 1.324→1.347 Å / Num. unique obs: 51 / CC1/2: 0.48 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5T35, 4QY4 Resolution: 1.32→26.4 Å / Cor.coef. Fo:Fc: 0.959 / Cor.coef. Fo:Fc free: 0.95 / SU R Cruickshank DPI: 0.073 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.076 / SU Rfree Blow DPI: 0.074 / SU Rfree Cruickshank DPI: 0.072
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 255.41 Å2 / Biso mean: 29.14 Å2 / Biso min: 10.72 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.19 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 1.32→26.4 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.32→1.35 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj