+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7z74 | ||||||
|---|---|---|---|---|---|---|---|
| Title | PI3KC2a core in complex with PITCOIN2 | ||||||
 Components Components | Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha | ||||||
 Keywords Keywords |  TRANSFERASE / PI3KC2 alpha / TRANSFERASE / PI3KC2 alpha /  kinase kinase | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of zinc ion transmembrane transport / Synthesis of PIPs at the early endosome membrane / Synthesis of PIPs at the late endosome membrane / Synthesis of PIPs at the Golgi membrane / Synthesis of PIPs at the plasma membrane / autophagosome organization / Golgi Associated Vesicle Biogenesis /  phosphatidylinositol-4-phosphate 3-kinase / phosphatidylinositol-4-phosphate 3-kinase /  Clathrin-mediated endocytosis / Clathrin-mediated endocytosis /  phosphatidylinositol 3-kinase complex ...negative regulation of zinc ion transmembrane transport / Synthesis of PIPs at the early endosome membrane / Synthesis of PIPs at the late endosome membrane / Synthesis of PIPs at the Golgi membrane / Synthesis of PIPs at the plasma membrane / autophagosome organization / Golgi Associated Vesicle Biogenesis / phosphatidylinositol 3-kinase complex ...negative regulation of zinc ion transmembrane transport / Synthesis of PIPs at the early endosome membrane / Synthesis of PIPs at the late endosome membrane / Synthesis of PIPs at the Golgi membrane / Synthesis of PIPs at the plasma membrane / autophagosome organization / Golgi Associated Vesicle Biogenesis /  phosphatidylinositol-4-phosphate 3-kinase / phosphatidylinositol-4-phosphate 3-kinase /  Clathrin-mediated endocytosis / Clathrin-mediated endocytosis /  phosphatidylinositol 3-kinase complex / 1-phosphatidylinositol-4-phosphate 3-kinase activity / 1-phosphatidylinositol-4,5-bisphosphate 3-kinase activity / phosphatidylinositol 3-kinase complex / 1-phosphatidylinositol-4-phosphate 3-kinase activity / 1-phosphatidylinositol-4,5-bisphosphate 3-kinase activity /  phosphatidylinositol-4,5-bisphosphate 3-kinase / phosphatidylinositol-4,5-bisphosphate 3-kinase /  clathrin-coated vesicle / clathrin-coated vesicle /  phosphatidylinositol 3-kinase / phosphatidylinositol-3-phosphate biosynthetic process / phosphatidylinositol 3-kinase / phosphatidylinositol-3-phosphate biosynthetic process /  clathrin binding / 1-phosphatidylinositol-3-kinase activity / positive regulation of cell migration involved in sprouting angiogenesis / clathrin binding / 1-phosphatidylinositol-3-kinase activity / positive regulation of cell migration involved in sprouting angiogenesis /  exocytosis / positive regulation of autophagy / cellular response to starvation / exocytosis / positive regulation of autophagy / cellular response to starvation /  phosphatidylinositol binding / phosphatidylinositol 3-kinase/protein kinase B signal transduction / phosphatidylinositol binding / phosphatidylinositol 3-kinase/protein kinase B signal transduction /  macroautophagy / macroautophagy /  trans-Golgi network / trans-Golgi network /  endocytosis / vesicle / endocytosis / vesicle /  phosphorylation / intracellular membrane-bounded organelle / phosphorylation / intracellular membrane-bounded organelle /  nucleoplasm / nucleoplasm /  ATP binding / ATP binding /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.5 Å MOLECULAR REPLACEMENT / Resolution: 2.5 Å | ||||||
 Authors Authors | Lo, W.T. / Roske, Y. / Daumke, O. / Haucke, V. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: Nat.Chem.Biol. / Year: 2023 Journal: Nat.Chem.Biol. / Year: 2023Title: Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha. Authors: Lo, W.T. / Belabed, H. / Kucukdisli, M. / Metag, J. / Roske, Y. / Prokofeva, P. / Ohashi, Y. / Horatscheck, A. / Cirillo, D. / Krauss, M. / Schmied, C. / Neuenschwander, M. / von Kries, J.P. ...Authors: Lo, W.T. / Belabed, H. / Kucukdisli, M. / Metag, J. / Roske, Y. / Prokofeva, P. / Ohashi, Y. / Horatscheck, A. / Cirillo, D. / Krauss, M. / Schmied, C. / Neuenschwander, M. / von Kries, J.P. / Medard, G. / Kuster, B. / Perisic, O. / Williams, R.L. / Daumke, O. / Payrastre, B. / Severin, S. / Nazare, M. / Haucke, V. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7z74.cif.gz 7z74.cif.gz | 349.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7z74.ent.gz pdb7z74.ent.gz | 282.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7z74.json.gz 7z74.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/z7/7z74 https://data.pdbj.org/pub/pdb/validation_reports/z7/7z74 ftp://data.pdbj.org/pub/pdb/validation_reports/z7/7z74 ftp://data.pdbj.org/pub/pdb/validation_reports/z7/7z74 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7z75C  8a9iC  7bi4S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 102486.023 Da / Num. of mol.: 1 / Mutation: 533-544/GSGS;550-665/SGAGSGA; F735A;F736A;L809A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Gene: Pik3c2a, Cpk / Production host: Mus musculus (house mouse) / Gene: Pik3c2a, Cpk / Production host:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm)References: UniProt: Q61194,  phosphatidylinositol 3-kinase, phosphatidylinositol 3-kinase,  phosphatidylinositol-4,5-bisphosphate 3-kinase, phosphatidylinositol-4,5-bisphosphate 3-kinase,  phosphatidylinositol-4-phosphate 3-kinase phosphatidylinositol-4-phosphate 3-kinase | ||||
|---|---|---|---|---|---|
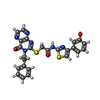
| #2: Chemical | ChemComp-IKC / ~{ | ||||
| #3: Chemical | ChemComp-EDO /  Ethylene glycol Ethylene glycol#4: Water | ChemComp-HOH / |  Water WaterHas ligand of interest | Y | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.82 Å3/Da / Density % sol: 56.5 % |
|---|---|
Crystal grow | Temperature: 298 K / Method: vapor diffusion / pH: 7.5 / Details: 0.1MTris, 8-6% PEG3350, 0.2-0.1M MgSO4 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  BESSY BESSY  / Beamline: 14.1 / Wavelength: 0.9184 Å / Beamline: 14.1 / Wavelength: 0.9184 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Apr 7, 2021 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9184 Å / Relative weight: 1 : 0.9184 Å / Relative weight: 1 |
| Reflection | Resolution: 2.5→43.26 Å / Num. obs: 41035 / % possible obs: 99.67 % / Redundancy: 5.5 % / CC1/2: 0.998 / Net I/σ(I): 9.02 |
| Reflection shell | Resolution: 2.5→2.589 Å / Num. unique obs: 4007 / CC1/2: 0.172 / % possible all: 99.68 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 7BI4 Resolution: 2.5→43.26 Å / SU ML: 0.54 / Cross valid method: THROUGHOUT / σ(F): 1.33 / Phase error: 35.07 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 210.34 Å2 / Biso mean: 86.8764 Å2 / Biso min: 30 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.5→43.26 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 15
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj