[English] 日本語
 Yorodumi
Yorodumi- PDB-7xib: Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ub... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7xib | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-disordered form) | |||||||||||||||
 Components Components |
| |||||||||||||||
 Keywords Keywords | TRANSFERASE/DNA /  DNA methyltransferase / TRANSFERASE-DNA COMPLEX DNA methyltransferase / TRANSFERASE-DNA COMPLEX | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching / : / epigenetic programming of gene expression / cellular response to bisphenol A / negative regulation of vascular associated smooth muscle cell apoptotic process / DNA-methyltransferase activity /  DNA (cytosine-5-)-methyltransferase / DNA (cytosine-5-)-methyltransferase /  DNA (cytosine-5-)-methyltransferase activity / DNA methylation-dependent heterochromatin formation / SUMOylation of DNA methylation proteins ...negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching / : / epigenetic programming of gene expression / cellular response to bisphenol A / negative regulation of vascular associated smooth muscle cell apoptotic process / DNA-methyltransferase activity / DNA (cytosine-5-)-methyltransferase activity / DNA methylation-dependent heterochromatin formation / SUMOylation of DNA methylation proteins ...negative regulation of vascular associated smooth muscle cell differentiation involved in phenotypic switching / : / epigenetic programming of gene expression / cellular response to bisphenol A / negative regulation of vascular associated smooth muscle cell apoptotic process / DNA-methyltransferase activity /  DNA (cytosine-5-)-methyltransferase / DNA (cytosine-5-)-methyltransferase /  DNA (cytosine-5-)-methyltransferase activity / DNA methylation-dependent heterochromatin formation / SUMOylation of DNA methylation proteins / negative regulation of gene expression via chromosomal CpG island methylation / female germ cell nucleus / methyl-CpG binding / pericentric heterochromatin / positive regulation of vascular associated smooth muscle cell proliferation / DNA (cytosine-5-)-methyltransferase activity / DNA methylation-dependent heterochromatin formation / SUMOylation of DNA methylation proteins / negative regulation of gene expression via chromosomal CpG island methylation / female germ cell nucleus / methyl-CpG binding / pericentric heterochromatin / positive regulation of vascular associated smooth muscle cell proliferation /  DNA methylation / DNA methylation /  replication fork / PRC2 methylates histones and DNA / Defective pyroptosis / promoter-specific chromatin binding / cellular response to amino acid stimulus / NoRC negatively regulates rRNA expression / negative regulation of gene expression / DNA-templated transcription / positive regulation of gene expression / negative regulation of transcription by RNA polymerase II / replication fork / PRC2 methylates histones and DNA / Defective pyroptosis / promoter-specific chromatin binding / cellular response to amino acid stimulus / NoRC negatively regulates rRNA expression / negative regulation of gene expression / DNA-templated transcription / positive regulation of gene expression / negative regulation of transcription by RNA polymerase II /  DNA binding / DNA binding /  RNA binding / zinc ion binding / RNA binding / zinc ion binding /  nucleoplasm / nucleoplasm /  nucleus nucleusSimilarity search - Function | |||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human)synthetic construct (others) | |||||||||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.23 Å cryo EM / Resolution: 2.23 Å | |||||||||||||||
 Authors Authors | Onoda, H. / Kikuchi, A. / Kori, S. / Yoshimi, S. / Yamagata, A. / Arita, K. | |||||||||||||||
| Funding support |  Japan, 4items Japan, 4items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for activation of DNMT1. Authors: Amika Kikuchi / Hiroki Onoda / Kosuke Yamaguchi / Satomi Kori / Shun Matsuzawa / Yoshie Chiba / Shota Tanimoto / Sae Yoshimi / Hiroki Sato / Atsushi Yamagata / Mikako Shirouzu / Naruhiko ...Authors: Amika Kikuchi / Hiroki Onoda / Kosuke Yamaguchi / Satomi Kori / Shun Matsuzawa / Yoshie Chiba / Shota Tanimoto / Sae Yoshimi / Hiroki Sato / Atsushi Yamagata / Mikako Shirouzu / Naruhiko Adachi / Jafar Sharif / Haruhiko Koseki / Atsuya Nishiyama / Makoto Nakanishi / Pierre-Antoine Defossez / Kyohei Arita /   Abstract: DNMT1 is an essential enzyme that maintains genomic DNA methylation, and its function is regulated by mechanisms that are not yet fully understood. Here, we report the cryo-EM structure of human ...DNMT1 is an essential enzyme that maintains genomic DNA methylation, and its function is regulated by mechanisms that are not yet fully understood. Here, we report the cryo-EM structure of human DNMT1 bound to its two natural activators: hemimethylated DNA and ubiquitinated histone H3. We find that a hitherto unstudied linker, between the RFTS and CXXC domains, plays a key role for activation. It contains a conserved α-helix which engages a crucial "Toggle" pocket, displacing a previously described inhibitory linker, and allowing the DNA Recognition Helix to spring into the active conformation. This is accompanied by large-scale reorganization of the inhibitory RFTS and CXXC domains, allowing the enzyme to gain full activity. Our results therefore provide a mechanistic basis for the activation of DNMT1, with consequences for basic research and drug design. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7xib.cif.gz 7xib.cif.gz | 187.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7xib.ent.gz pdb7xib.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  7xib.json.gz 7xib.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xi/7xib https://data.pdbj.org/pub/pdb/validation_reports/xi/7xib ftp://data.pdbj.org/pub/pdb/validation_reports/xi/7xib ftp://data.pdbj.org/pub/pdb/validation_reports/xi/7xib | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 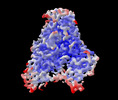 33201MC  7xi9C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 143106.000 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: DNMT1 / Plasmid: pFastBac / Cell line (production host): sf-9 / Production host: Homo sapiens (human) / Gene: DNMT1 / Plasmid: pFastBac / Cell line (production host): sf-9 / Production host:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HMReferences: UniProt: P26358,  DNA (cytosine-5-)-methyltransferase DNA (cytosine-5-)-methyltransferase | ||||
|---|---|---|---|---|---|
| #2: DNA chain | Mass: 3725.469 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) | ||||
| #3: DNA chain | Mass: 3679.464 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.) synthetic construct (others) | ||||
| #4: Chemical | | #5: Water | ChemComp-HOH / |  Water WaterHas ligand of interest | Y | |
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Value: 0.17 MDa / Experimental value: NO | ||||||||||||||||||||||||
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||
| Source (recombinant) | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM | ||||||||||||||||||||||||
| Buffer solution | pH: 7.5 | ||||||||||||||||||||||||
| Buffer component |
| ||||||||||||||||||||||||
| Specimen | Conc.: 0.5 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YESDetails: This sample was monodisperse by Size-exclusion chromatography | ||||||||||||||||||||||||
| Specimen support | Grid material: COPPER / Grid mesh size: 300 divisions/in. / Grid type: Quantifoil R1.2/1.3 | ||||||||||||||||||||||||
Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277 K / Details: Blot for 4 seconds before plunging |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1800 nm / Nominal defocus min: 800 nm / Cs Bright-field microscopy / Nominal defocus max: 1800 nm / Nominal defocus min: 800 nm / Cs : 2.7 mm / C2 aperture diameter: 50 µm : 2.7 mm / C2 aperture diameter: 50 µm |
| Specimen holder | Cryogen: NITROGEN |
| Image recording | Electron dose: 49 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Num. of grids imaged: 1 / Num. of real images: 4068 |
| Image scans | Width: 4092 / Height: 5760 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| |||||||||||||||||||||||||||||||||||||||||||||
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | |||||||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 3798046 | |||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry : C1 (asymmetric) : C1 (asymmetric) | |||||||||||||||||||||||||||||||||||||||||||||
3D reconstruction | Resolution: 2.23 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 897446 / Algorithm: EXACT BACK PROJECTION / Num. of class averages: 1 / Symmetry type: POINT | |||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: BACKBONE TRACE / Space: REAL | |||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building |
| |||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Cross valid method: NONE Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2 | |||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 40.28 Å2 | |||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller



 PDBj
PDBj










































