+ Open data
Open data
- Basic information
Basic information
| Entry | 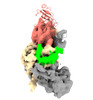 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of apo-DNMT1 (aa:351-1616) | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords |  DNA methyltransferase / DNA methyltransferase /  TRANSFERASE TRANSFERASE | |||||||||||||||
| Function / homology |  : :  Function and homology information Function and homology information | |||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
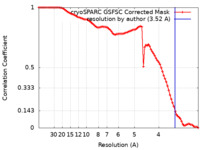
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.52 Å cryo EM / Resolution: 3.52 Å | |||||||||||||||
 Authors Authors | Onoda H / Kikuchi A / Kori S / Yoshimi S / Yamagata A / Matsuzawa S / Arita K | |||||||||||||||
| Funding support |  Japan, 4 items Japan, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for activation of DNMT1. Authors: Amika Kikuchi / Hiroki Onoda / Kosuke Yamaguchi / Satomi Kori / Shun Matsuzawa / Yoshie Chiba / Shota Tanimoto / Sae Yoshimi / Hiroki Sato / Atsushi Yamagata / Mikako Shirouzu / Naruhiko ...Authors: Amika Kikuchi / Hiroki Onoda / Kosuke Yamaguchi / Satomi Kori / Shun Matsuzawa / Yoshie Chiba / Shota Tanimoto / Sae Yoshimi / Hiroki Sato / Atsushi Yamagata / Mikako Shirouzu / Naruhiko Adachi / Jafar Sharif / Haruhiko Koseki / Atsuya Nishiyama / Makoto Nakanishi / Pierre-Antoine Defossez / Kyohei Arita /   Abstract: DNMT1 is an essential enzyme that maintains genomic DNA methylation, and its function is regulated by mechanisms that are not yet fully understood. Here, we report the cryo-EM structure of human ...DNMT1 is an essential enzyme that maintains genomic DNA methylation, and its function is regulated by mechanisms that are not yet fully understood. Here, we report the cryo-EM structure of human DNMT1 bound to its two natural activators: hemimethylated DNA and ubiquitinated histone H3. We find that a hitherto unstudied linker, between the RFTS and CXXC domains, plays a key role for activation. It contains a conserved α-helix which engages a crucial "Toggle" pocket, displacing a previously described inhibitory linker, and allowing the DNA Recognition Helix to spring into the active conformation. This is accompanied by large-scale reorganization of the inhibitory RFTS and CXXC domains, allowing the enzyme to gain full activity. Our results therefore provide a mechanistic basis for the activation of DNMT1, with consequences for basic research and drug design. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33298.map.gz emd_33298.map.gz | 32.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33298-v30.xml emd-33298-v30.xml emd-33298.xml emd-33298.xml | 23.8 KB 23.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33298_fsc.xml emd_33298_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_33298.png emd_33298.png | 70.8 KB | ||
| Masks |  emd_33298_msk_1.map emd_33298_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33298.cif.gz emd-33298.cif.gz | 7 KB | ||
| Others |  emd_33298_half_map_1.map.gz emd_33298_half_map_1.map.gz emd_33298_half_map_2.map.gz emd_33298_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33298 http://ftp.pdbj.org/pub/emdb/structures/EMD-33298 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33298 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33298 | HTTPS FTP |
-Related structure data
| Related structure data |  7xi9C  7xibC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33298.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33298.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.477 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33298_msk_1.map emd_33298_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33298_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33298_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human DNMT1, 351_1616 with H3Ub2
| Entire | Name: human DNMT1, 351_1616 with H3Ub2 |
|---|---|
| Components |
|
-Supramolecule #1: human DNMT1, 351_1616 with H3Ub2
| Supramolecule | Name: human DNMT1, 351_1616 with H3Ub2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 160 KDa |
-Supramolecule #2: DNA (cytosine-5)-methyltransferase 1
| Supramolecule | Name: DNA (cytosine-5)-methyltransferase 1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Histone H3.1
| Supramolecule | Name: Histone H3.1 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: H3Ub2
| Supramolecule | Name: H3Ub2 / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|
-Macromolecule #1: DNA (cytosine-5)-methyltransferase 1
| Macromolecule | Name: DNA (cytosine-5)-methyltransferase 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number:  DNA (cytosine-5-)-methyltransferase DNA (cytosine-5-)-methyltransferase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / Strain: Isoform 1 Homo sapiens (human) / Strain: Isoform 1 |
| Recombinant expression | Organism:   Baculovirus expression vector pFastBac1-HM Baculovirus expression vector pFastBac1-HM |
| Sequence | String: PKCIQCGQYL DDPDLKYGQH PPDAVDEPQ MLTNEKLSIF D ANESGFES YEALPQHKLT CF SVYCKHG HLCPIDTGLI EKN IELFFS GSAKPIYDDD PSLE GGVNG KNLGPINEWW ITGFD GGEK ALIGFSTSFA EYILMD PSP EYAPIFGLMQ EKIYISK IV ...String: PKCIQCGQYL DDPDLKYGQH PPDAVDEPQ MLTNEKLSIF D ANESGFES YEALPQHKLT CF SVYCKHG HLCPIDTGLI EKN IELFFS GSAKPIYDDD PSLE GGVNG KNLGPINEWW ITGFD GGEK ALIGFSTSFA EYILMD PSP EYAPIFGLMQ EKIYISK IV VEFLQSNSDS TYEDLINK I ETTVPPSGLN LNRFTEDSL LRHAQFVVEQ VESYDEAGDS DEQPIFLTP CMRDLIKLAG V TLGQRRAQ ARRQTIRHST RE KDRGPTK ATTTKLVYQI FDT FFAEQI EKDDREDKEN AFKR RRCGV CEVCQQPECG KCKAC KDMV KFGGSGRSKQ ACQERR CPN MAMKEADDDE EVDDNIP EM PSPKKMHQGK KKKQNKNR I SWVGEAVKTD GKKSYYKKV CIDAETLEVG DCVSVIPDDS SKPLYLARV TALWEDSSNG Q MFHAHWFC AGTDTVLGAT SD PLELFLV DECEDMQLSY IHS KVKVIY KAPSENWAME GGMD PESLL EGDDGKTYFY QLWYD QDYA RFESPPKTQP TEDNKF KFC VSCARLAEMR QKEIPRV LE QLEDLDSRVL YYSATKNG I LYRVGDGVYL PPEAFTFNI KLSSPVKRPR KEPVDEDLYP EHYRKYSDY IKGSNLDAPE P YRIGRIKE IFCPKKSNGR PN ETDIKIR VNKFYRPENT HKS TPASYH ADINLLYWSD EEAV VDFKA VQGRCTVEYG EDLPE CVQV YSMGGPNRFY FLEAYN AKS KSFEDPPNHA RSPGNKG KG KGKGKGKPKS QACEPSEP E IEIKLPKLRT LDVFSGCGG LSEGFHQAGI SDTLWAIEMW DPAAQAFRL NNPGSTVFTE D CNILLKLV MAGETTNSRG QR LPQKGDV EMLCGGPPCQ GFS GMNRFN SRTYSKFKNS LVVS FLSYC DYYRPRFFLL ENVRN FVSF KRSMVLKLTL RCLVRM GYQ CTFGVLQAGQ YGVAQTR RR AIILAAAPGE KLPLFPEP L HVFAPRACQL SVVVDDKKF VSNITRLSSG PFRTITVRDT MSDLPEVRN GASALEISYN G EPQSWFQR QLRGAQYQPI LR DHICKDM SALVAARMRH IPL APGSDW RDLPNIEVRL SDGT MARKL RYTHHDRKNG RSSSG ALRG VCSCVEAGKA CDPAAR QFN TLIPWCLPHT GNRHNHW AG LYGRLEWDGF FSTTVTNP E PMGKQGRVLH PEQHRVVSV RECARSQGFP DTYRLFGNIL DKHRQVGNA VPPPLAKAIG L EIKLCMLA KARESASAKI KE EEAAKD UniProtKB:  UNIPROTKB: NP_001370.1 UNIPROTKB: NP_001370.1 |
-Macromolecule #2: Histone H3.1 K14R/K27R/K36R/K37W
| Macromolecule | Name: Histone H3.1 K14R/K27R/K36R/K37W / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: ARTKQTARK STGGRAPRKQ LATKAARRS A PATGGVRW |
-Macromolecule #3: Ubiquitin
| Macromolecule | Name: Ubiquitin / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGKQL EDGRTLSDYN IQKESTLHL VLRLRGG |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 4 seconds before plunging. | |||||||||||||||
| Details | This sample was monodisperse by Size-exclusion chromatography |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.4000000000000001 µm / Nominal defocus min: 0.8 µm Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 1.4000000000000001 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 1 / Number real images: 1869 / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: B / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller




 Z
Z Y
Y X
X