+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7x9d | ||||||
|---|---|---|---|---|---|---|---|
| Title | DNMT3B in complex with harmine | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  TRANSFERASE / Inhibitor / Comple / TRANSFERASE / Inhibitor / Comple /  Methyltransferase Methyltransferase | ||||||
| Function / homology |  Function and homology information Function and homology informationretrotransposon silencing by heterochromatin formation / : / epigenetic programing of female pronucleus / chorionic trophoblast cell differentiation / DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates / DNA-methyltransferase activity /  genomic imprinting / genomic imprinting /  DNA (cytosine-5-)-methyltransferase / DNA (cytosine-5-)-methyltransferase /  DNA (cytosine-5-)-methyltransferase activity / autosome genomic imprinting ...retrotransposon silencing by heterochromatin formation / : / epigenetic programing of female pronucleus / chorionic trophoblast cell differentiation / DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates / DNA-methyltransferase activity / DNA (cytosine-5-)-methyltransferase activity / autosome genomic imprinting ...retrotransposon silencing by heterochromatin formation / : / epigenetic programing of female pronucleus / chorionic trophoblast cell differentiation / DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates / DNA-methyltransferase activity /  genomic imprinting / genomic imprinting /  DNA (cytosine-5-)-methyltransferase / DNA (cytosine-5-)-methyltransferase /  DNA (cytosine-5-)-methyltransferase activity / autosome genomic imprinting / negative regulation of DNA methylation-dependent heterochromatin formation / SUMOylation of DNA methylation proteins / : / DNA methylation-dependent heterochromatin formation / ESC/E(Z) complex / DNA metabolic process / negative regulation of gene expression, epigenetic / male meiosis I / DNA (cytosine-5-)-methyltransferase activity / autosome genomic imprinting / negative regulation of DNA methylation-dependent heterochromatin formation / SUMOylation of DNA methylation proteins / : / DNA methylation-dependent heterochromatin formation / ESC/E(Z) complex / DNA metabolic process / negative regulation of gene expression, epigenetic / male meiosis I /  catalytic complex / catalytic complex /  heterochromatin / heterochromatin /  enzyme activator activity / enzyme activator activity /  DNA methylation / PRC2 methylates histones and DNA / condensed nuclear chromosome / Defective pyroptosis / DNA methylation / PRC2 methylates histones and DNA / condensed nuclear chromosome / Defective pyroptosis /  stem cell differentiation / placenta development / NoRC negatively regulates rRNA expression / transcription corepressor activity / stem cell differentiation / placenta development / NoRC negatively regulates rRNA expression / transcription corepressor activity /  methylation / methylation /  spermatogenesis / positive regulation of gene expression / spermatogenesis / positive regulation of gene expression /  enzyme binding / negative regulation of transcription by RNA polymerase II / enzyme binding / negative regulation of transcription by RNA polymerase II /  DNA binding / DNA binding /  nucleoplasm / nucleoplasm /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.08 Å MOLECULAR REPLACEMENT / Resolution: 3.08 Å | ||||||
 Authors Authors | Cho, C.-C. / Yuan, H.S. | ||||||
| Funding support |  Taiwan, 1items Taiwan, 1items
| ||||||
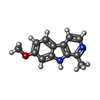
 Citation Citation |  Journal: Acs Chem.Biol. / Year: 2023 Journal: Acs Chem.Biol. / Year: 2023Title: Mechanistic Insights into Harmine-Mediated Inhibition of Human DNA Methyltransferases and Prostate Cancer Cell Growth. Authors: Cho, C.C. / Lin, C.J. / Huang, H.H. / Yang, W.Z. / Fei, C.Y. / Lin, H.Y. / Lee, M.S. / Yuan, H.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7x9d.cif.gz 7x9d.cif.gz | 200.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7x9d.ent.gz pdb7x9d.ent.gz | 159 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7x9d.json.gz 7x9d.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/x9/7x9d https://data.pdbj.org/pub/pdb/validation_reports/x9/7x9d ftp://data.pdbj.org/pub/pdb/validation_reports/x9/7x9d ftp://data.pdbj.org/pub/pdb/validation_reports/x9/7x9d | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6kdpS S: Starting model for refinement |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 32715.859 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: DNMT3B / Production host: Homo sapiens (human) / Gene: DNMT3B / Production host:   Escherichia coli (E. coli) Escherichia coli (E. coli)References: UniProt: Q9UBC3,  DNA (cytosine-5-)-methyltransferase DNA (cytosine-5-)-methyltransferase#2: Protein | Mass: 23627.107 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: DNMT3L / Production host: Homo sapiens (human) / Gene: DNMT3L / Production host:   Escherichia coli (E. coli) / References: UniProt: Q9UJW3 Escherichia coli (E. coli) / References: UniProt: Q9UJW3#3: Chemical |  Harmine HarmineHas ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.51 Å3/Da / Density % sol: 72.71 % |
|---|---|
Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / Details: bis-tris propane pH 7.0, sodium formate |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSRRC NSRRC  / Beamline: BL15A1 / Wavelength: 1 Å / Beamline: BL15A1 / Wavelength: 1 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RAYONIX MX-300 / Detector: CCD / Date: Mar 16, 2021 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 3.08→30 Å / Num. obs: 35934 / % possible obs: 97.7 % / Redundancy: 5.2 % / Biso Wilson estimate: 67.42 Å2 / Rmerge(I) obs: 0.084 / Rpim(I) all: 0.042 / Rrim(I) all: 0.095 / Χ2: 0.886 / Net I/σ(I): 8 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6KDP Resolution: 3.08→29.94 Å / SU ML: 0.34 / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 26.59 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 157.7 Å2 / Biso mean: 62.7657 Å2 / Biso min: 18.34 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3.08→29.94 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 14
|
 Movie
Movie Controller
Controller



 PDBj
PDBj