+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7vlr | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of SUR2B in complex with Mg-ATP/ADP | ||||||||||||
 Components Components | Isoform SUR2B of ATP-binding cassette sub-family C member 9 | ||||||||||||
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  SUR2B / SUR2B /  ABC transporter ABC transporter | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsubstrate-dependent cell migration, cell contraction /  ATP sensitive Potassium channels / ABC-family proteins mediated transport / ATP sensitive Potassium channels / ABC-family proteins mediated transport /  circulatory system development / inward rectifying potassium channel / circulatory system development / inward rectifying potassium channel /  sulfonylurea receptor activity / cardiac conduction / ATPase-coupled monoatomic cation transmembrane transporter activity / coronary vasculature development / cardiac muscle cell contraction ...substrate-dependent cell migration, cell contraction / sulfonylurea receptor activity / cardiac conduction / ATPase-coupled monoatomic cation transmembrane transporter activity / coronary vasculature development / cardiac muscle cell contraction ...substrate-dependent cell migration, cell contraction /  ATP sensitive Potassium channels / ABC-family proteins mediated transport / ATP sensitive Potassium channels / ABC-family proteins mediated transport /  circulatory system development / inward rectifying potassium channel / circulatory system development / inward rectifying potassium channel /  sulfonylurea receptor activity / cardiac conduction / ATPase-coupled monoatomic cation transmembrane transporter activity / coronary vasculature development / cardiac muscle cell contraction / inorganic cation transmembrane transport / sulfonylurea receptor activity / cardiac conduction / ATPase-coupled monoatomic cation transmembrane transporter activity / coronary vasculature development / cardiac muscle cell contraction / inorganic cation transmembrane transport /  syntaxin binding / syntaxin binding /  action potential / Ion homeostasis / response to ATP / action potential / Ion homeostasis / response to ATP /  heterocyclic compound binding / potassium ion import across plasma membrane / blood vessel development / monoatomic cation transmembrane transport / ATPase-coupled transmembrane transporter activity / potassium channel regulator activity / ABC-type transporter activity / heart morphogenesis / potassium ion transmembrane transport / heterocyclic compound binding / potassium ion import across plasma membrane / blood vessel development / monoatomic cation transmembrane transport / ATPase-coupled transmembrane transporter activity / potassium channel regulator activity / ABC-type transporter activity / heart morphogenesis / potassium ion transmembrane transport /  T-tubule / negative regulation of blood pressure / T-tubule / negative regulation of blood pressure /  sarcomere / acrosomal vesicle / transmembrane transport / potassium ion transport / sarcomere / acrosomal vesicle / transmembrane transport / potassium ion transport /  regulation of blood pressure / fibroblast proliferation / defense response to virus / transmembrane transporter binding / response to xenobiotic stimulus / regulation of blood pressure / fibroblast proliferation / defense response to virus / transmembrane transporter binding / response to xenobiotic stimulus /  ATP hydrolysis activity / ATP hydrolysis activity /  ATP binding / ATP binding /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||||||||
| Biological species |   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) | ||||||||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.4 Å cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Chen, L. / Ding, D. | ||||||||||||
| Funding support |  China, 3items China, 3items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural identification of vasodilator binding sites on the SUR2 subunit. Authors: Dian Ding / Jing-Xiang Wu / Xinli Duan / Songling Ma / Lipeng Lai / Lei Chen /  Abstract: ATP-sensitive potassium channels (K), composed of Kir6 and SUR subunits, convert the metabolic status of the cell into electrical signals. Pharmacological activation of SUR2- containing K channels by ...ATP-sensitive potassium channels (K), composed of Kir6 and SUR subunits, convert the metabolic status of the cell into electrical signals. Pharmacological activation of SUR2- containing K channels by class of small molecule drugs known as K openers leads to hyperpolarization of excitable cells and to vasodilation. Thus, K openers could be used to treat cardiovascular diseases. However, where these vasodilators bind to K and how they activate the channel remains elusive. Here, we present cryo-EM structures of SUR2A and SUR2B subunits in complex with Mg-nucleotides and P1075 or levcromakalim, two chemically distinct K openers that are specific to SUR2. Both P1075 and levcromakalim bind to a common site in the transmembrane domain (TMD) of the SUR2 subunit, which is between TMD1 and TMD2 and is embraced by TM10, TM11, TM12, TM14, and TM17. These K openers synergize with Mg-nucleotides to stabilize SUR2 in the NBD-dimerized occluded state to activate the channel. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7vlr.cif.gz 7vlr.cif.gz | 237 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7vlr.ent.gz pdb7vlr.ent.gz | 186.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7vlr.json.gz 7vlr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vl/7vlr https://data.pdbj.org/pub/pdb/validation_reports/vl/7vlr ftp://data.pdbj.org/pub/pdb/validation_reports/vl/7vlr ftp://data.pdbj.org/pub/pdb/validation_reports/vl/7vlr | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 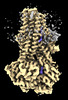 32024MC  7vlsC  7vltC  7vluC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 174488.562 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Rattus norvegicus (Norway rat) / Gene: Abcc9, Sur2 / Production host: Rattus norvegicus (Norway rat) / Gene: Abcc9, Sur2 / Production host:   Homo sapiens (human) / References: UniProt: Q63563 Homo sapiens (human) / References: UniProt: Q63563 | ||||
|---|---|---|---|---|---|
| #2: Chemical | ChemComp-Y01 / | ||||
| #3: Chemical | ChemComp-ADP /  Adenosine diphosphate Adenosine diphosphate | ||||
| #4: Chemical | | #5: Chemical | ChemComp-ATP / |  Adenosine triphosphate Adenosine triphosphateHas ligand of interest | Y | |
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: ATP-binding cassette sub-family C member 9, isoform B / Type: COMPLEX / Entity ID: #1 / Source: RECOMBINANT |
|---|---|
| Molecular weight | Value: 174 kDa/nm / Experimental value: NO |
| Source (natural) | Organism:   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) |
| Source (recombinant) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1800 nm / Nominal defocus min: 1500 nm Bright-field microscopy / Nominal defocus max: 1800 nm / Nominal defocus min: 1500 nm |
| Image recording | Electron dose: 52 e/Å2 / Detector mode: SUPER-RESOLUTION / Film or detector model: GATAN K2 SUMMIT (4k x 4k) |
- Processing
Processing
| EM software | Name: cryoSPARC / Version: 3.1.0 / Category: 3D reconstruction |
|---|---|
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION |
| Symmetry | Point symmetry : C1 (asymmetric) : C1 (asymmetric) |
3D reconstruction | Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 36180 / Symmetry type: POINT |
 Movie
Movie Controller
Controller






 PDBj
PDBj











