[English] 日本語
 Yorodumi
Yorodumi- PDB-7uzp: parathyroid hormone 1 receptor extracellular domain complexed wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7uzp | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | parathyroid hormone 1 receptor extracellular domain complexed with a peptide ligand containing three beta-amino acids | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  SIGNALING PROTEIN / SIGNALING PROTEIN /  parathyroid hormone 1 receptor / parathyroid hormone 1 receptor /  signaling / beta-amino acid signaling / beta-amino acid | |||||||||
| Function / homology |  Function and homology information Function and homology information parathyroid hormone receptor activity / G protein-coupled peptide receptor activity / Class B/2 (Secretin family receptors) / osteoblast development / positive regulation of inositol phosphate biosynthetic process / parathyroid hormone receptor activity / G protein-coupled peptide receptor activity / Class B/2 (Secretin family receptors) / osteoblast development / positive regulation of inositol phosphate biosynthetic process /  bone mineralization / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / bone mineralization / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger /  peptide hormone binding / chondrocyte differentiation / cell maturation ... peptide hormone binding / chondrocyte differentiation / cell maturation ... parathyroid hormone receptor activity / G protein-coupled peptide receptor activity / Class B/2 (Secretin family receptors) / osteoblast development / positive regulation of inositol phosphate biosynthetic process / parathyroid hormone receptor activity / G protein-coupled peptide receptor activity / Class B/2 (Secretin family receptors) / osteoblast development / positive regulation of inositol phosphate biosynthetic process /  bone mineralization / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / bone mineralization / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger /  peptide hormone binding / chondrocyte differentiation / cell maturation / peptide hormone binding / chondrocyte differentiation / cell maturation /  bone resorption / bone resorption /  skeletal system development / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / intracellular calcium ion homeostasis / : / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (s) signalling events / basolateral plasma membrane / in utero embryonic development / cell population proliferation / cell surface receptor signaling pathway / skeletal system development / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / intracellular calcium ion homeostasis / : / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (s) signalling events / basolateral plasma membrane / in utero embryonic development / cell population proliferation / cell surface receptor signaling pathway /  receptor complex / apical plasma membrane / G protein-coupled receptor signaling pathway / negative regulation of cell population proliferation / positive regulation of cell population proliferation / protein homodimerization activity / receptor complex / apical plasma membrane / G protein-coupled receptor signaling pathway / negative regulation of cell population proliferation / positive regulation of cell population proliferation / protein homodimerization activity /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.29 Å MOLECULAR REPLACEMENT / Resolution: 2.29 Å | |||||||||
 Authors Authors | Yu, Z. / Bruchs, A.T. / Bingman, C.A. / Gellman, S.H. | |||||||||
| Funding support |  United States, 1items United States, 1items
| |||||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2022 Journal: Proc.Natl.Acad.Sci.USA / Year: 2022Title: Altered signaling at the PTH receptor via modified agonist contacts with the extracellular domain provides a path to prolonged agonism in vivo. Authors: Liu, S. / Yu, Z. / Daley, E.J. / Bingman, C.A. / Bruchs, A.T. / Gardella, T.J. / Gellman, S.H. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7uzp.cif.gz 7uzp.cif.gz | 234.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7uzp.ent.gz pdb7uzp.ent.gz | 192.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7uzp.json.gz 7uzp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uz/7uzp https://data.pdbj.org/pub/pdb/validation_reports/uz/7uzp ftp://data.pdbj.org/pub/pdb/validation_reports/uz/7uzp ftp://data.pdbj.org/pub/pdb/validation_reports/uz/7uzp | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 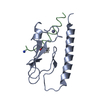 7uzoC  6fj3S S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 11995.670 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: PTH1R, PTHR, PTHR1 / Production host: Homo sapiens (human) / Gene: PTH1R, PTHR, PTHR1 / Production host:   Escherichia coli (E. coli) / References: UniProt: Q03431 Escherichia coli (E. coli) / References: UniProt: Q03431#2: Protein/peptide | Mass: 2779.161 Da / Num. of mol.: 3 / Source method: obtained synthetically / Source: (synth.)   Homo sapiens (human) Homo sapiens (human)#3: Chemical |  Ethylene glycol Ethylene glycol#4: Chemical | ChemComp-ZN / | #5: Water | ChemComp-HOH / |  Water WaterHas ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.34 Å3/Da / Density % sol: 47.51 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop Details: 28 % v/v PEG Smear Low, 0.1 M tris-HCl pH 8, 0.15 M Sodium citrate, 1% ethylene glycol, 0.4 mM ZnSO4 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 21-ID-G / Wavelength: 0.97856 Å / Beamline: 21-ID-G / Wavelength: 0.97856 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: RAYONIX MX-300 / Detector: CCD / Date: Jun 19, 2021 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 0.97856 Å / Relative weight: 1 : 0.97856 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.29→34.54 Å / Num. obs: 18565 / % possible obs: 99.7 % / Redundancy: 7.501 % / Biso Wilson estimate: 41.05 Å2 / CC1/2: 0.997 / Rmerge(I) obs: 0.1 / Rrim(I) all: 0.107 / Χ2: 0.873 / Net I/σ(I): 12.62 / Num. measured all: 139252 / Scaling rejects: 24 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6FJ3 Resolution: 2.29→34.54 Å / SU ML: 0.33 / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 27.47 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.1 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 224.55 Å2 / Biso mean: 60.6824 Å2 / Biso min: 22.37 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.29→34.54 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 7
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller


 PDBj
PDBj




