[English] 日本語
 Yorodumi
Yorodumi- PDB-7g97: ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in c... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7g97 | ||||||
|---|---|---|---|---|---|---|---|
| Title | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z57899718 | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  CYTOSOLIC PROTEIN / SGC - Diamond I04-1 fragment screening / PanDDA / XChemExplorer CYTOSOLIC PROTEIN / SGC - Diamond I04-1 fragment screening / PanDDA / XChemExplorer | ||||||
| Function / homology |  Function and homology information Function and homology informationasymmetric neuroblast division / negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress / aortic valve formation / alpha-beta T cell lineage commitment / mitotic cleavage furrow formation / bone trabecula morphogenesis / positive regulation of lipase activity / endothelial tube lumen extension / skeletal muscle satellite cell migration / positive regulation of vascular associated smooth muscle contraction ...asymmetric neuroblast division / negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress / aortic valve formation / alpha-beta T cell lineage commitment / mitotic cleavage furrow formation / bone trabecula morphogenesis / positive regulation of lipase activity / endothelial tube lumen extension / skeletal muscle satellite cell migration / positive regulation of vascular associated smooth muscle contraction / angiotensin-mediated vasoconstriction involved in regulation of systemic arterial blood pressure / SLIT2:ROBO1 increases RHOA activity / RHO GTPases Activate Rhotekin and Rhophilins / Roundabout signaling pathway / negative regulation of intracellular steroid hormone receptor signaling pathway / cellular response to muramyl dipeptide / Axonal growth inhibition (RHOA activation) / Axonal growth stimulation / regulation of neural precursor cell proliferation / cleavage furrow formation / positive regulation of neuron migration / regulation of modification of postsynaptic actin cytoskeleton / regulation of osteoblast proliferation / forebrain radial glial cell differentiation / negative regulation of microtubule depolymerization /  cell junction assembly / apical junction assembly / regulation of systemic arterial blood pressure by endothelin / regulation of Rho protein signal transduction / cellular hyperosmotic response / cellular response to chemokine / negative regulation of cell migration involved in sprouting angiogenesis / negative regulation of necroptotic process / beta selection / establishment of epithelial cell apical/basal polarity / regulation of modification of postsynaptic structure / negative regulation of cell size / RHO GTPases Activate ROCKs / negative regulation of oxidative phosphorylation / negative regulation of motor neuron apoptotic process / RHO GTPases activate CIT / PCP/CE pathway / Sema4D induced cell migration and growth-cone collapse / RHO GTPases activate KTN1 / apolipoprotein A-I-mediated signaling pathway / positive regulation of podosome assembly / negative regulation of cell-substrate adhesion / cell junction assembly / apical junction assembly / regulation of systemic arterial blood pressure by endothelin / regulation of Rho protein signal transduction / cellular hyperosmotic response / cellular response to chemokine / negative regulation of cell migration involved in sprouting angiogenesis / negative regulation of necroptotic process / beta selection / establishment of epithelial cell apical/basal polarity / regulation of modification of postsynaptic structure / negative regulation of cell size / RHO GTPases Activate ROCKs / negative regulation of oxidative phosphorylation / negative regulation of motor neuron apoptotic process / RHO GTPases activate CIT / PCP/CE pathway / Sema4D induced cell migration and growth-cone collapse / RHO GTPases activate KTN1 / apolipoprotein A-I-mediated signaling pathway / positive regulation of podosome assembly / negative regulation of cell-substrate adhesion /  Wnt signaling pathway, planar cell polarity pathway / Sema4D mediated inhibition of cell attachment and migration / regulation of small GTPase mediated signal transduction / ossification involved in bone maturation / positive regulation of alpha-beta T cell differentiation / Wnt signaling pathway, planar cell polarity pathway / Sema4D mediated inhibition of cell attachment and migration / regulation of small GTPase mediated signal transduction / ossification involved in bone maturation / positive regulation of alpha-beta T cell differentiation /  odontogenesis / motor neuron apoptotic process / PI3K/AKT activation / odontogenesis / motor neuron apoptotic process / PI3K/AKT activation /  wound healing, spreading of cells / positive regulation of leukocyte adhesion to vascular endothelial cell / apical junction complex / wound healing, spreading of cells / positive regulation of leukocyte adhesion to vascular endothelial cell / apical junction complex /  regulation of focal adhesion assembly / negative chemotaxis / RHOB GTPase cycle / regulation of focal adhesion assembly / negative chemotaxis / RHOB GTPase cycle /  myosin binding / NRAGE signals death through JNK / EPHA-mediated growth cone collapse / myosin binding / NRAGE signals death through JNK / EPHA-mediated growth cone collapse /  stress fiber assembly / regulation of neuron projection development / RHOC GTPase cycle / androgen receptor signaling pathway / positive regulation of cytokinesis / cerebral cortex cell migration / ERBB2 Regulates Cell Motility / cellular response to cytokine stimulus / stress fiber assembly / regulation of neuron projection development / RHOC GTPase cycle / androgen receptor signaling pathway / positive regulation of cytokinesis / cerebral cortex cell migration / ERBB2 Regulates Cell Motility / cellular response to cytokine stimulus /  cleavage furrow / semaphorin-plexin signaling pathway / Rho protein signal transduction / ficolin-1-rich granule membrane / cleavage furrow / semaphorin-plexin signaling pathway / Rho protein signal transduction / ficolin-1-rich granule membrane /  mitotic spindle assembly / RHOA GTPase cycle / endothelial cell migration / bicellular tight junction / positive regulation of T cell migration / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / regulation of microtubule cytoskeleton organization / cytoplasmic microtubule organization / skeletal muscle tissue development / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors / mitotic spindle assembly / RHOA GTPase cycle / endothelial cell migration / bicellular tight junction / positive regulation of T cell migration / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / regulation of microtubule cytoskeleton organization / cytoplasmic microtubule organization / skeletal muscle tissue development / negative regulation of extrinsic apoptotic signaling pathway via death domain receptors /  regulation of cell migration / negative regulation of reactive oxygen species biosynthetic process / RHO GTPases activate PKNs / positive regulation of stress fiber assembly / GPVI-mediated activation cascade / EPHB-mediated forward signaling / substantia nigra development / positive regulation of neuron differentiation / substrate adhesion-dependent cell spreading / cell-matrix adhesion / guanyl-nucleotide exchange factor activity / regulation of cell migration / negative regulation of reactive oxygen species biosynthetic process / RHO GTPases activate PKNs / positive regulation of stress fiber assembly / GPVI-mediated activation cascade / EPHB-mediated forward signaling / substantia nigra development / positive regulation of neuron differentiation / substrate adhesion-dependent cell spreading / cell-matrix adhesion / guanyl-nucleotide exchange factor activity /  small monomeric GTPase / G protein activity / secretory granule membrane small monomeric GTPase / G protein activity / secretory granule membraneSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  FOURIER SYNTHESIS / FOURIER SYNTHESIS /  molecular replacement / Resolution: 2.3 Å molecular replacement / Resolution: 2.3 Å | ||||||
 Authors Authors | Bradshaw, W.J. / Katis, V.L. / Bountra, C. / von Delft, F. / Brennan, P.E. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: ARHGEF2 PanDDA analysis group deposition Authors: Bradshaw, W.J. / Katis, V.L. / Bountra, C. / von Delft, F. / Brennan, P.E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7g97.cif.gz 7g97.cif.gz | 99.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7g97.ent.gz pdb7g97.ent.gz | 75.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7g97.json.gz 7g97.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/g9/7g97 https://data.pdbj.org/pub/pdb/validation_reports/g9/7g97 ftp://data.pdbj.org/pub/pdb/validation_reports/g9/7g97 ftp://data.pdbj.org/pub/pdb/validation_reports/g9/7g97 | HTTPS FTP |
|---|
-Group deposition
| ID | G_1002269 (56 entries) |
|---|---|
| Title | ARHGEF2 PanDDA analysis group deposition |
| Type | changed state |
| Description | XDomainX of XOrganismX RHOA screened against the XXX Fragment Library by X-ray Crystallography at the XChem facility of Diamond Light Source beamline I04-1 |
-Related structure data
| Related structure data |  8bntS S: Starting model for refinement |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 2 types, 2 molecules AB
| #1: Protein |  / Rho cDNA clone 12 / h12 / Rho cDNA clone 12 / h12Mass: 20925.975 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: RHOA, ARH12, ARHA, RHO12 / Production host: Homo sapiens (human) / Gene: RHOA, ARH12, ARHA, RHO12 / Production host:   Escherichia coli (E. coli) / References: UniProt: P61586, Escherichia coli (E. coli) / References: UniProt: P61586,  small monomeric GTPase small monomeric GTPase |
|---|---|
| #2: Protein | Mass: 28644.150 Da / Num. of mol.: 1 / Mutation: None Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: ARHGEF2, KIAA0651, LFP40 / Production host: Homo sapiens (human) / Gene: ARHGEF2, KIAA0651, LFP40 / Production host:   Escherichia coli (E. coli) / References: UniProt: Q92974 Escherichia coli (E. coli) / References: UniProt: Q92974 |
-Non-polymers , 4 types, 92 molecules 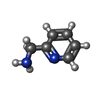






| #3: Chemical | ChemComp-APY /  2-Picolylamine 2-Picolylamine | ||||
|---|---|---|---|---|---|
| #4: Chemical |  Dimethyl sulfoxide Dimethyl sulfoxide#5: Chemical | ChemComp-FMT / |  Formic acid Formic acid#6: Water | ChemComp-HOH / |  Water Water |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.51 Å3/Da / Density % sol: 50.96 % / Mosaicity: 0 ° |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 8.5 / Details: 100 mM Tris, 2.6M sodium formate |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Diamond Diamond  / Beamline: I04-1 / Wavelength: 0.92124 Å / Beamline: I04-1 / Wavelength: 0.92124 Å | |||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Mar 2, 2023 | |||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Scattering type: x-ray | |||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 0.92124 Å / Relative weight: 1 : 0.92124 Å / Relative weight: 1 | |||||||||||||||||||||||||||
| Reflection | Resolution: 2.3→71.27 Å / Num. obs: 23427 / % possible obs: 100 % / Redundancy: 21 % / CC1/2: 0.992 / Rmerge(I) obs: 0.636 / Rpim(I) all: 0.143 / Rrim(I) all: 0.666 / Net I/σ(I): 6.1 / Num. measured all: 491371 / Scaling rejects: 9 | |||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1 / % possible all: 100
|
-Phasing
Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  FOURIER SYNTHESIS FOURIER SYNTHESISStarting model: 8BNT Resolution: 2.3→66.94 Å / Cor.coef. Fo:Fc: 0.954 / Cor.coef. Fo:Fc free: 0.921 / SU B: 8.687 / SU ML: 0.198 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R: 0.285 / ESU R Free: 0.23 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : REFINED INDIVIDUALLY
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 129.91 Å2 / Biso mean: 36.521 Å2 / Biso min: 15.68 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.3→66.94 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.3→2.358 Å / Total num. of bins used: 20
|
 Movie
Movie Controller
Controller












 PDBj
PDBj
















