[English] 日本語
 Yorodumi
Yorodumi- PDB-6xs8: Crystal structure of Chaetomium thermophilum Vps29 complexed with... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6xs8 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Chaetomium thermophilum Vps29 complexed with RaPID-derived cyclic peptide RT-D3 | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PROTEIN TRANSPORT/INHIBITOR /  Vps29 / Vps29 /  Retromer / Retromer /  Endosome / Endosome /  Protein transport / Protein transport /  cyclic peptide / cyclic peptide /  inhibitor / PROTEIN TRANSPORT-INHIBITOR complex inhibitor / PROTEIN TRANSPORT-INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology information retromer complex / retromer complex /  retrograde transport, endosome to Golgi / retrograde transport, endosome to Golgi /  protein transport / protein transport /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus)synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 1.95009854987 Å molecular replacement / Resolution: 1.95009854987 Å | ||||||
 Authors Authors | Chen, K.-E. / Guo, Q. / Collins, B.M. | ||||||
| Funding support |  Australia, 1items Australia, 1items
| ||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: De novo macrocyclic peptides for inhibiting, stabilizing, and probing the function of the retromer endosomal trafficking complex. Authors: Kai-En Chen / Qian Guo / Timothy A Hill / Yi Cui / Amy K Kendall / Zhe Yang / Ryan J Hall / Michael D Healy / Joanna Sacharz / Suzanne J Norwood / Sachini Fonseka / Boyang Xie / Robert C ...Authors: Kai-En Chen / Qian Guo / Timothy A Hill / Yi Cui / Amy K Kendall / Zhe Yang / Ryan J Hall / Michael D Healy / Joanna Sacharz / Suzanne J Norwood / Sachini Fonseka / Boyang Xie / Robert C Reid / Natalya Leneva / Robert G Parton / Rajesh Ghai / David A Stroud / David P Fairlie / Hiroaki Suga / Lauren P Jackson / Rohan D Teasdale / Toby Passioura / Brett M Collins /    Abstract: The retromer complex (Vps35-Vps26-Vps29) is essential for endosomal membrane trafficking and signaling. Mutation of the retromer subunit Vps35 causes late-onset Parkinson’s disease, while viral and ...The retromer complex (Vps35-Vps26-Vps29) is essential for endosomal membrane trafficking and signaling. Mutation of the retromer subunit Vps35 causes late-onset Parkinson’s disease, while viral and bacterial pathogens can hijack the complex during cellular infection. To modulate and probe its function, we have created a novel series of macrocyclic peptides that bind retromer with high affinity and specificity. Crystal structures show that most of the cyclic peptides bind to Vps29 via a Pro-Leu–containing sequence, structurally mimicking known interactors such as TBC1D5 and blocking their interaction with retromer in vitro and in cells. By contrast, macrocyclic peptide RT-L4 binds retromer at the Vps35-Vps26 interface and is a more effective molecular chaperone than reported small molecules, suggesting a new therapeutic avenue for targeting retromer. Last, tagged peptides can be used to probe the cellular localization of retromer and its functional interactions in cells, providing novel tools for studying retromer function. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6xs8.cif.gz 6xs8.cif.gz | 62.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6xs8.ent.gz pdb6xs8.ent.gz | 41.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6xs8.json.gz 6xs8.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xs/6xs8 https://data.pdbj.org/pub/pdb/validation_reports/xs/6xs8 ftp://data.pdbj.org/pub/pdb/validation_reports/xs/6xs8 ftp://data.pdbj.org/pub/pdb/validation_reports/xs/6xs8 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6xs5C  6xs7C  6xs9C  6xsaC  5w8mS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 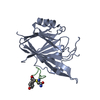
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||
| Unit cell |
| ||||||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein |  Vacuole VacuoleMass: 22387.654 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Chaetomium thermophilum (fungus) / Production host: Chaetomium thermophilum (fungus) / Production host:   Escherichia coli BL21(DE3) (bacteria) / References: UniProt: G0RZB5 Escherichia coli BL21(DE3) (bacteria) / References: UniProt: G0RZB5 |
|---|---|
| #2: Protein/peptide | Mass: 1567.783 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Cyclic peptide RT-D3 / Source: (synth.) synthetic construct (others) |
| #3: Water | ChemComp-HOH /  Water Water |
| Has ligand of interest | Y |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.33 Å3/Da / Density % sol: 47.13 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / Details: 0.18 M ammonium citrate dibasic and 20% PEG 3350 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Australian Synchrotron Australian Synchrotron  / Beamline: MX2 / Wavelength: 0.9537 Å / Beamline: MX2 / Wavelength: 0.9537 Å | ||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Nov 10, 2018 | ||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 0.9537 Å / Relative weight: 1 : 0.9537 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||
| Reflection | Resolution: 1.83→42.75 Å / Num. obs: 20527 / % possible obs: 99.6 % / Redundancy: 10.2 % / Biso Wilson estimate: 16.885 Å2 / CC1/2: 1 / Rmerge(I) obs: 0.039 / Rpim(I) all: 0.013 / Rrim(I) all: 0.041 / Net I/σ(I): 28 / Num. measured all: 209530 / Scaling rejects: 3 | ||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR | Model details: Phaser MODE: MR_AUTO
|
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5W8M Resolution: 1.95009854987→41.1509291116 Å / SU ML: 0.199634091036 / Cross valid method: FREE R-VALUE / σ(F): 0 / Phase error: 22.266463367 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| |||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 21.446980057 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.95009854987→41.1509291116 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj

