[English] 日本語
 Yorodumi
Yorodumi- PDB-6wc5: Crystal Structure of a Ternary MEF2B/NKX2-5/myocardin enhancer DN... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6wc5 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of a Ternary MEF2B/NKX2-5/myocardin enhancer DNA Complex | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  TRANSCRIPTION / TRANSCRIPTION /  transcription complex / Cardiogenesis / transcription complex / Cardiogenesis /  Carcinogenesis / MADS-box and Homeobox Carcinogenesis / MADS-box and Homeobox | ||||||
| Function / homology |  Function and homology information Function and homology informationNkx-2.5 complex / Purkinje myocyte differentiation / right ventricular cardiac muscle tissue morphogenesis / septum secundum development / proepicardium development / pulmonary myocardium development / cardiac ventricle formation / apoptotic process involved in heart morphogenesis / atrioventricular node cell fate commitment / bundle of His development ...Nkx-2.5 complex / Purkinje myocyte differentiation / right ventricular cardiac muscle tissue morphogenesis / septum secundum development / proepicardium development / pulmonary myocardium development / cardiac ventricle formation / apoptotic process involved in heart morphogenesis / atrioventricular node cell fate commitment / bundle of His development / atrial cardiac muscle cell development / ventricular cardiac myofibril assembly / atrioventricular node cell development / embryonic heart tube left/right pattern formation / positive regulation of cardioblast differentiation / atrial cardiac muscle tissue development / atrioventricular node development / ventricular cardiac muscle cell development / regulation of cardiac muscle cell proliferation / atrial septum morphogenesis / positive regulation of heart contraction / Physiological factors / negative regulation of myotube differentiation /  cardiac conduction system development / YAP1- and WWTR1 (TAZ)-stimulated gene expression / pharyngeal system development / positive regulation of sodium ion transport / negative regulation of epithelial cell apoptotic process / outflow tract septum morphogenesis / cardiac muscle tissue morphogenesis / ventricular trabecula myocardium morphogenesis / heart trabecula formation / adult heart development / embryonic heart tube development / aortic valve morphogenesis / cardiac muscle cell development / Cardiogenesis / negative regulation of cardiac muscle cell apoptotic process / muscle organ development / cardiac muscle cell proliferation / epithelial cell apoptotic process / DNA-binding transcription activator activity / ventricular septum morphogenesis / heart looping / cardiac conduction system development / YAP1- and WWTR1 (TAZ)-stimulated gene expression / pharyngeal system development / positive regulation of sodium ion transport / negative regulation of epithelial cell apoptotic process / outflow tract septum morphogenesis / cardiac muscle tissue morphogenesis / ventricular trabecula myocardium morphogenesis / heart trabecula formation / adult heart development / embryonic heart tube development / aortic valve morphogenesis / cardiac muscle cell development / Cardiogenesis / negative regulation of cardiac muscle cell apoptotic process / muscle organ development / cardiac muscle cell proliferation / epithelial cell apoptotic process / DNA-binding transcription activator activity / ventricular septum morphogenesis / heart looping /  Myogenesis / cardiac septum morphogenesis / thyroid gland development / Myogenesis / cardiac septum morphogenesis / thyroid gland development /  hemopoiesis / regulation of cardiac conduction / positive regulation of transcription initiation by RNA polymerase II / regulation of cardiac muscle contraction / hemopoiesis / regulation of cardiac conduction / positive regulation of transcription initiation by RNA polymerase II / regulation of cardiac muscle contraction /  vasculogenesis / heart morphogenesis / cardiac muscle contraction / spleen development / epithelial cell differentiation / positive regulation of neuron differentiation / epithelial cell proliferation / positive regulation of epithelial cell proliferation / vasculogenesis / heart morphogenesis / cardiac muscle contraction / spleen development / epithelial cell differentiation / positive regulation of neuron differentiation / epithelial cell proliferation / positive regulation of epithelial cell proliferation /  protein-DNA complex / negative regulation of canonical Wnt signaling pathway / protein-DNA complex / negative regulation of canonical Wnt signaling pathway /  histone deacetylase binding / RNA polymerase II transcription regulator complex / sequence-specific double-stranded DNA binding / histone deacetylase binding / RNA polymerase II transcription regulator complex / sequence-specific double-stranded DNA binding /  cell junction / cell junction /  heart development / DNA-binding transcription activator activity, RNA polymerase II-specific / heart development / DNA-binding transcription activator activity, RNA polymerase II-specific /  transcription regulator complex / RNA polymerase II-specific DNA-binding transcription factor binding / sequence-specific DNA binding / transcription by RNA polymerase II / transcription regulator complex / RNA polymerase II-specific DNA-binding transcription factor binding / sequence-specific DNA binding / transcription by RNA polymerase II /  cell differentiation / cell differentiation /  protein dimerization activity / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of DNA-templated transcription / protein dimerization activity / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of DNA-templated transcription /  chromatin binding / positive regulation of cell population proliferation / chromatin binding / positive regulation of cell population proliferation /  chromatin / positive regulation of gene expression / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / protein-containing complex / chromatin / positive regulation of gene expression / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / protein-containing complex /  DNA binding / DNA binding /  nucleoplasm / nucleoplasm /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human)synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.9 Å MOLECULAR REPLACEMENT / Resolution: 2.9 Å | ||||||
 Authors Authors | Chen, L. / Lei, X. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2020 Journal: J.Mol.Biol. / Year: 2020Title: Crystal Structures of Ternary Complexes of MEF2 and NKX2-5 Bound to DNA Reveal a Disease Related Protein-Protein Interaction Interface. Authors: Lei, X. / Zhao, J. / Sagendorf, J.M. / Rajashekar, N. / Xu, J. / Dantas Machado, A.C. / Sen, C. / Rohs, R. / Feng, P. / Chen, L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6wc5.cif.gz 6wc5.cif.gz | 305.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6wc5.ent.gz pdb6wc5.ent.gz | 243.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6wc5.json.gz 6wc5.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/wc/6wc5 https://data.pdbj.org/pub/pdb/validation_reports/wc/6wc5 ftp://data.pdbj.org/pub/pdb/validation_reports/wc/6wc5 ftp://data.pdbj.org/pub/pdb/validation_reports/wc/6wc5 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6wc2C  1n6jS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 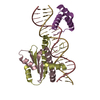
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 0 / Refine code: 0
|
 Movie
Movie Controller
Controller







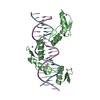




 PDBj
PDBj






































