+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6pll | ||||||
|---|---|---|---|---|---|---|---|
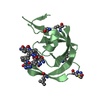
| Title | Crystal structure of the ZIG-8 IG1 homodimer | ||||||
 Components Components | Zwei Ig domain protein zig-8 | ||||||
 Keywords Keywords |  CELL ADHESION / CELL ADHESION /  Cell Surface Receptor / Cell Surface Receptor /  Immunoglobulin Superfamily / Immunoglobulin Superfamily /  Nervous System / Nervous System /  Homodimer Homodimer | ||||||
| Function / homology |  Function and homology information Function and homology informationneuron projection membrane / synapse organization / extracellular region Similarity search - Function | ||||||
| Biological species |   Caenorhabditis elegans (invertebrata) Caenorhabditis elegans (invertebrata) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.693 Å MOLECULAR REPLACEMENT / Resolution: 2.693 Å | ||||||
 Authors Authors | Cheng, S. / Ozkan, E. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Proc. Natl. Acad. Sci. U.S.A. / Year: 2019 Journal: Proc. Natl. Acad. Sci. U.S.A. / Year: 2019Title: Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence. Authors: Cheng, S. / Park, Y. / Kurleto, J.D. / Jeon, M. / Zinn, K. / Thornton, J.W. / Ozkan, E. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6pll.cif.gz 6pll.cif.gz | 157.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6pll.ent.gz pdb6pll.ent.gz | 124.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6pll.json.gz 6pll.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/pl/6pll https://data.pdbj.org/pub/pdb/validation_reports/pl/6pll ftp://data.pdbj.org/pub/pdb/validation_reports/pl/6pll ftp://data.pdbj.org/pub/pdb/validation_reports/pl/6pll | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6on6C  6on9SC  6onbC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 14297.955 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Caenorhabditis elegans (invertebrata) / Gene: zig-8, Y39E4B.8 / Cell line (production host): High Five cells / Production host: Caenorhabditis elegans (invertebrata) / Gene: zig-8, Y39E4B.8 / Cell line (production host): High Five cells / Production host:   Trichoplusia ni (cabbage looper) / References: UniProt: G5ED00 Trichoplusia ni (cabbage looper) / References: UniProt: G5ED00#2: Chemical |  Sulfate Sulfate#3: Sugar | ChemComp-NAG / |  N-Acetylglucosamine N-Acetylglucosamine#4: Water | ChemComp-HOH / |  Water WaterHas ligand of interest | N | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.46 Å3/Da / Density % sol: 50.08 % |
|---|---|
Crystal grow | Temperature: 294 K / Method: vapor diffusion, sitting drop Details: 0.2 M Ammonium dihydrogen phosphate, 20% (w/v) PEG 3350 |
-Data collection
| Diffraction | Mean temperature: 120 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-BM / Wavelength: 0.9791928 Å / Beamline: 19-BM / Wavelength: 0.9791928 Å |
| Detector | Type: ADSC QUANTUM 210r / Detector: CCD / Date: Apr 13, 2019 |
| Radiation | Monochromator: Si (111) double-crystal monochromator / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9791928 Å / Relative weight: 1 : 0.9791928 Å / Relative weight: 1 |
| Reflection | Resolution: 2.69→50 Å / Num. obs: 12889 / % possible obs: 99.7 % / Observed criterion σ(I): -3 / Redundancy: 21.4 % / CC1/2: 0.998 / Rmerge(I) obs: 0.225 / Rrim(I) all: 0.23 / Net I/σ(I): 15.33 |
| Reflection shell | Resolution: 2.69→2.86 Å / Redundancy: 17.7 % / Rmerge(I) obs: 2.597 / Mean I/σ(I) obs: 1.05 / Num. unique obs: 1967 / CC1/2: 0.529 / Rrim(I) all: 2.673 / % possible all: 98.3 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6ON9 Resolution: 2.693→46.381 Å / SU ML: 0.47 / Cross valid method: FREE R-VALUE / σ(F): 0 / Phase error: 28.63
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.693→46.381 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj








