+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 6nhg | ||||||
|---|---|---|---|---|---|---|---|
| タイトル | Rhodobacter sphaeroides Mitochondrial respiratory chain complex | ||||||
 要素 要素 |
| ||||||
 キーワード キーワード |  OXIDOREDUCTASE (酸化還元酵素) / OXIDOREDUCTASE (酸化還元酵素) /  Mitochondrial respiratory chain complex (電子伝達系) / Mitochondrial respiratory chain complex (電子伝達系) /  cytochrome bc1 (ユビキノール-シトクロムcレダクターゼ) / inhibitors / cytochrome bc1 (ユビキノール-シトクロムcレダクターゼ) / inhibitors /  electron transfer (電子移動反応) electron transfer (電子移動反応) | ||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Respiratory electron transport / mitochondrial respiratory chain complex III / quinol-cytochrome-c reductase /  ubiquinol-cytochrome-c reductase activity / ubiquinol-cytochrome-c reductase activity /  Mitochondrial protein degradation / mitochondrial electron transport, ubiquinol to cytochrome c / Mitochondrial protein degradation / mitochondrial electron transport, ubiquinol to cytochrome c /  ubiquinone binding / ubiquinone binding /  respirasome / respiratory electron transport chain / respirasome / respiratory electron transport chain /  ミトコンドリア ...Respiratory electron transport / mitochondrial respiratory chain complex III / quinol-cytochrome-c reductase / ミトコンドリア ...Respiratory electron transport / mitochondrial respiratory chain complex III / quinol-cytochrome-c reductase /  ubiquinol-cytochrome-c reductase activity / ubiquinol-cytochrome-c reductase activity /  Mitochondrial protein degradation / mitochondrial electron transport, ubiquinol to cytochrome c / Mitochondrial protein degradation / mitochondrial electron transport, ubiquinol to cytochrome c /  ubiquinone binding / ubiquinone binding /  respirasome / respiratory electron transport chain / respirasome / respiratory electron transport chain /  ミトコンドリア / 2 iron, 2 sulfur cluster binding / ミトコンドリア / 2 iron, 2 sulfur cluster binding /  metalloendopeptidase activity / metalloendopeptidase activity /  ミトコンドリア内膜 / ミトコンドリア内膜 /  oxidoreductase activity / oxidoreductase activity /  heme binding / heme binding /  ミトコンドリア / ミトコンドリア /  タンパク質分解 / タンパク質分解 /  生体膜 / 生体膜 /  metal ion binding / metal ion binding /  細胞膜 細胞膜類似検索 - 分子機能 | ||||||
| 生物種 |   Bos taurus (ウシ) Bos taurus (ウシ) | ||||||
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 2.8 Å 分子置換 / 解像度: 2.8 Å | ||||||
 データ登録者 データ登録者 | Xia, D. / Zhou, F. / Esser, L. | ||||||
 引用 引用 |  ジャーナル: J.Biol.Chem. / 年: 2019 ジャーナル: J.Biol.Chem. / 年: 2019タイトル: Crystal structure of bacterial cytochromebc1in complex with azoxystrobin reveals a conformational switch of the Rieske iron-sulfur protein subunit. 著者: Esser, L. / Zhou, F. / Yu, C.A. / Xia, D. | ||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  6nhg.cif.gz 6nhg.cif.gz | 1.2 MB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb6nhg.ent.gz pdb6nhg.ent.gz | 1 MB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  6nhg.json.gz 6nhg.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/nh/6nhg https://data.pdbj.org/pub/pdb/validation_reports/nh/6nhg ftp://data.pdbj.org/pub/pdb/validation_reports/nh/6nhg ftp://data.pdbj.org/pub/pdb/validation_reports/nh/6nhg | HTTPS FTP |
|---|
-関連構造データ
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 単位格子 |
|
- 要素
要素
-Cytochrome b-c1 complex subunit ... , 9種, 9分子 ABEFGHIJK
| #1: タンパク質 | 分子量: 49266.254 Da / 分子数: 1 / 由来タイプ: 天然 / 由来: (天然)   Bos taurus (ウシ) / 参照: UniProt: P31800 Bos taurus (ウシ) / 参照: UniProt: P31800 |
|---|---|
| #2: タンパク質 | 分子量: 46575.469 Da / 分子数: 1 / 由来タイプ: 天然 / 由来: (天然)   Bos taurus (ウシ) / 参照: UniProt: P23004 Bos taurus (ウシ) / 参照: UniProt: P23004 |
| #5: タンパク質 | 分子量: 21640.580 Da / 分子数: 1 / 由来タイプ: 天然 / 由来: (天然)   Bos taurus (ウシ) / 参照: UniProt: P13272, quinol-cytochrome-c reductase Bos taurus (ウシ) / 参照: UniProt: P13272, quinol-cytochrome-c reductase |
| #6: タンパク質 | 分子量: 13370.206 Da / 分子数: 1 / 由来タイプ: 天然 / 由来: (天然)   Bos taurus (ウシ) / 参照: UniProt: P00129 Bos taurus (ウシ) / 参照: UniProt: P00129 |
| #7: タンパク質 | 分子量: 9448.833 Da / 分子数: 1 / 由来タイプ: 天然 / 由来: (天然)   Bos taurus (ウシ) / 参照: UniProt: P13271 Bos taurus (ウシ) / 参照: UniProt: P13271 |
| #8: タンパク質 | 分子量: 9189.116 Da / 分子数: 1 / 由来タイプ: 天然 / 由来: (天然)   Bos taurus (ウシ) / 参照: UniProt: P00126 Bos taurus (ウシ) / 参照: UniProt: P00126 |
| #9: タンパク質 | 分子量: 7964.259 Da / 分子数: 1 / 由来タイプ: 組換発現 詳細: Amino terminal signal peptide of mitiochondrial bc1 Rieske protein 由来: (組換発現)   Bos taurus (ウシ) / 遺伝子: UQCRFS1 / 発現宿主: Bos taurus (ウシ) / 遺伝子: UQCRFS1 / 発現宿主:   Bos taurus (ウシ) / 参照: UniProt: P13272, quinol-cytochrome-c reductase Bos taurus (ウシ) / 参照: UniProt: P13272, quinol-cytochrome-c reductase |
| #10: タンパク質 | 分子量: 7338.425 Da / 分子数: 1 / 由来タイプ: 天然 / 由来: (天然)   Bos taurus (ウシ) / 参照: UniProt: P00130 Bos taurus (ウシ) / 参照: UniProt: P00130 |
| #11: タンパク質 | 分子量: 6396.408 Da / 分子数: 1 / 由来タイプ: 天然 / 由来: (天然)   Bos taurus (ウシ) / 参照: UniProt: P07552 Bos taurus (ウシ) / 参照: UniProt: P07552 |
-タンパク質 , 2種, 2分子 CD
| #3: タンパク質 |  シトクロムb / Complex III subunit 3 / Complex III subunit III / Cytochrome b-c1 complex subunit 3 / Ubiquinol- ...Complex III subunit 3 / Complex III subunit III / Cytochrome b-c1 complex subunit 3 / Ubiquinol-cytochrome-c reductase complex cytochrome b subunit シトクロムb / Complex III subunit 3 / Complex III subunit III / Cytochrome b-c1 complex subunit 3 / Ubiquinol- ...Complex III subunit 3 / Complex III subunit III / Cytochrome b-c1 complex subunit 3 / Ubiquinol-cytochrome-c reductase complex cytochrome b subunit分子量: 42620.340 Da / 分子数: 1 / 由来タイプ: 天然 / 由来: (天然)   Bos taurus (ウシ) / 参照: UniProt: P00157 Bos taurus (ウシ) / 参照: UniProt: P00157 |
|---|---|
| #4: タンパク質 |  / Complex III subunit 4 / Complex III subunit IV / Cytochrome b-c1 complex subunit 4 / Ubiquinol- ...Complex III subunit 4 / Complex III subunit IV / Cytochrome b-c1 complex subunit 4 / Ubiquinol-cytochrome-c reductase complex cytochrome c1 subunit / Cytochrome c-1 / Complex III subunit 4 / Complex III subunit IV / Cytochrome b-c1 complex subunit 4 / Ubiquinol- ...Complex III subunit 4 / Complex III subunit IV / Cytochrome b-c1 complex subunit 4 / Ubiquinol-cytochrome-c reductase complex cytochrome c1 subunit / Cytochrome c-1分子量: 27323.277 Da / 分子数: 1 / 由来タイプ: 組換発現 / 由来: (組換発現)   Bos taurus (ウシ) / 遺伝子: CYC1 / 発現宿主: Bos taurus (ウシ) / 遺伝子: CYC1 / 発現宿主:   Bos taurus (ウシ) / 参照: UniProt: P00125 Bos taurus (ウシ) / 参照: UniProt: P00125 |
-非ポリマー , 9種, 46分子 


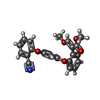












| #12: 化合物 | | #13: 化合物 |  Cardiolipin Cardiolipin#14: 化合物 |  Heme B Heme B#15: 化合物 | ChemComp-AZO / |  アゾキシストロビン アゾキシストロビン#16: 化合物 | ChemComp-8PE / ( | #17: 化合物 | ChemComp-HEC / |  Heme C Heme C#18: 化合物 | ChemComp-FES / |  鉄・硫黄クラスター 鉄・硫黄クラスター#19: 化合物 | ChemComp-MC3 / | #20: 水 | ChemComp-HOH / |  水 水 |
|---|
-実験情報
-実験
| 実験 | 手法:  X線回折 / 使用した結晶の数: 1 X線回折 / 使用した結晶の数: 1 |
|---|
- 試料調製
試料調製
| 結晶 | マシュー密度: 3.69 Å3/Da / 溶媒含有率: 66.63 % |
|---|---|
結晶化 | 温度: 277 K / 手法: 蒸気拡散法, シッティングドロップ法 / pH: 7.2 詳細: 50 mM MOPS 7.2 pH, 20 mM Ammonium acetate, 20% glycerol. Incubation with 2-5 mol excess of Azoxystrobin. Precipitant 12% PEG4000, 0.5 M KCl, 0.1% DHPC; Protein:PPT ratio 1:0.57 Temp details: constant |
-データ収集
| 回折 | 平均測定温度: 100 K / Serial crystal experiment: N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 放射光源 | 由来:  シンクロトロン / サイト: シンクロトロン / サイト:  APS APS  / ビームライン: 5ID-B / 波長: 1.739 Å / ビームライン: 5ID-B / 波長: 1.739 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 検出器 | タイプ: MAR CCD 165 mm / 検出器: CCD / 日付: 2001年2月3日 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 放射 | モノクロメーター: Si(111) / プロトコル: SINGLE WAVELENGTH / 単色(M)・ラウエ(L): M / 散乱光タイプ: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 放射波長 | 波長 : 1.739 Å / 相対比: 1 : 1.739 Å / 相対比: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 反射 | 解像度: 2.8→30 Å / Num. obs: 167324 / % possible obs: 99.8 % / 冗長度: 23.2 % / Rmerge(I) obs: 0.104 / Rpim(I) all: 0.022 / Rrim(I) all: 0.107 / Χ2: 1.009 / Net I/σ(I): 10 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 反射 シェル | Diffraction-ID: 1
|
- 解析
解析
| ソフトウェア |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 精密化 | 構造決定の手法 : :  分子置換 分子置換開始モデル: 1SQX 解像度: 2.8→29.87 Å / SU ML: 0.41 / 交差検証法: THROUGHOUT / σ(F): 1.33 / 位相誤差: 32.44
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 溶媒の処理 | 減衰半径: 0.9 Å / VDWプローブ半径: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 原子変位パラメータ | Biso max: 270.33 Å2 / Biso mean: 108.8564 Å2 / Biso min: 37.99 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化ステップ | サイクル: final / 解像度: 2.8→29.87 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS精密化 シェル | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 27
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化 TLS | 手法: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 精密化 TLSグループ |
|
 ムービー
ムービー コントローラー
コントローラー

















 PDBj
PDBj




















