[English] 日本語
 Yorodumi
Yorodumi- PDB-6ln2: Crystal structure of full length human GLP1 receptor in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ln2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of full length human GLP1 receptor in complex with Fab fragment (Fab7F38) | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  MEMBRANE PROTEIN / Full length Human GLP1 receptor / Class B / Fab7F38 / TMD / NAM PF06372222 / LCP MEMBRANE PROTEIN / Full length Human GLP1 receptor / Class B / Fab7F38 / TMD / NAM PF06372222 / LCP | ||||||
| Function / homology |  Function and homology information Function and homology informationalkane catabolic process /  glucagon-like peptide 1 receptor activity / glucagon-like peptide 1 receptor activity /  glucagon receptor activity / hormone secretion / positive regulation of blood pressure / glucagon receptor activity / hormone secretion / positive regulation of blood pressure /  post-translational protein targeting to membrane, translocation / post-translational protein targeting to membrane, translocation /  regulation of heart contraction / response to psychosocial stress / regulation of heart contraction / response to psychosocial stress /  peptide hormone binding / activation of adenylate cyclase activity ...alkane catabolic process / peptide hormone binding / activation of adenylate cyclase activity ...alkane catabolic process /  glucagon-like peptide 1 receptor activity / glucagon-like peptide 1 receptor activity /  glucagon receptor activity / hormone secretion / positive regulation of blood pressure / glucagon receptor activity / hormone secretion / positive regulation of blood pressure /  post-translational protein targeting to membrane, translocation / post-translational protein targeting to membrane, translocation /  regulation of heart contraction / response to psychosocial stress / regulation of heart contraction / response to psychosocial stress /  peptide hormone binding / activation of adenylate cyclase activity / negative regulation of blood pressure / cAMP-mediated signaling / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Glucagon-type ligand receptors / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / transmembrane signaling receptor activity / positive regulation of cytosolic calcium ion concentration / G alpha (s) signalling events / peptide hormone binding / activation of adenylate cyclase activity / negative regulation of blood pressure / cAMP-mediated signaling / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Glucagon-type ligand receptors / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / transmembrane signaling receptor activity / positive regulation of cytosolic calcium ion concentration / G alpha (s) signalling events /  electron transfer activity / learning or memory / cell surface receptor signaling pathway / iron ion binding / electron transfer activity / learning or memory / cell surface receptor signaling pathway / iron ion binding /  membrane / membrane /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human)  Clostridium pasteurianum (bacteria) Clostridium pasteurianum (bacteria)  Mus musculus (house mouse) Mus musculus (house mouse) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.2 Å MOLECULAR REPLACEMENT / Resolution: 3.2 Å | ||||||
 Authors Authors | Wu, F. / Yang, L. / Hang, K. / Laursen, M. / Wu, L. / Han, G.W. / Ren, Q. / Roed, N.K. / Lin, G. / Hanson, M. ...Wu, F. / Yang, L. / Hang, K. / Laursen, M. / Wu, L. / Han, G.W. / Ren, Q. / Roed, N.K. / Lin, G. / Hanson, M. / Jiang, H. / Wang, M. / Reedtz-Runge, S. / Song, G. / Stevens, R.C. | ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Full-length human GLP-1 receptor structure without orthosteric ligands. Authors: Wu, F. / Yang, L. / Hang, K. / Laursen, M. / Wu, L. / Han, G.W. / Ren, Q. / Roed, N.K. / Lin, G. / Hanson, M.A. / Jiang, H. / Wang, M.W. / Reedtz-Runge, S. / Song, G. / Stevens, R.C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ln2.cif.gz 6ln2.cif.gz | 351.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ln2.ent.gz pdb6ln2.ent.gz | 282.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ln2.json.gz 6ln2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ln/6ln2 https://data.pdbj.org/pub/pdb/validation_reports/ln/6ln2 ftp://data.pdbj.org/pub/pdb/validation_reports/ln/6ln2 ftp://data.pdbj.org/pub/pdb/validation_reports/ln/6ln2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5nx2S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | AUTHORS STATE THAT THE BIOLOGICAL UNIT IS UNKNOWN. |
- Components
Components
-Antibody , 2 types, 2 molecules BC
| #2: Antibody | Mass: 23307.939 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Production host: Mus musculus (house mouse) / Production host:   Mus musculus (house mouse) Mus musculus (house mouse) |
|---|---|
| #3: Antibody | Mass: 24476.268 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Production host: Mus musculus (house mouse) / Production host:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Protein / Sugars , 2 types, 2 molecules A

| #1: Protein | Mass: 54619.996 Da / Num. of mol.: 1 Mutation: S193C,I196F,S225A,M233C,S271A,I317C,G318I,K346A,C347F,G361C,E387D Source method: isolated from a genetically manipulated source Details: The fusion protein of GLP1 receptor (UNP residues 24-260), Rubredoxin (UNP residues 1-54) and GLP1 receptor (UNP residues 262-439) Source: (gene. exp.)   Homo sapiens (human), (gene. exp.) Homo sapiens (human), (gene. exp.)   Clostridium pasteurianum (bacteria) Clostridium pasteurianum (bacteria)Gene: GLP1R / Production host:   Spodoptera frugiperda (fall armyworm) / References: UniProt: P43220, UniProt: P00268 Spodoptera frugiperda (fall armyworm) / References: UniProt: P43220, UniProt: P00268 |
|---|---|
| #4: Sugar | ChemComp-NAG /  N-Acetylglucosamine N-Acetylglucosamine |
-Non-polymers , 2 types, 2 molecules 
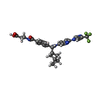

| #5: Chemical | ChemComp-ZN / |
|---|---|
| #6: Chemical | ChemComp-97Y / |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.21 Å3/Da / Density % sol: 61.66 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: lipidic cubic phase Details: 200-300 mM Ammonium formate, 36% PEG 400, 5%-10% (w/v) Guanidine hydrochloride PH range: pH 6.2-6.6 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SPring-8 SPring-8  / Beamline: BL45XU / Wavelength: 1 Å / Beamline: BL45XU / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: May 15, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 3.2→45.7 Å / Num. obs: 22019 / % possible obs: 97.8 % / Redundancy: 4.8 % / CC1/2: 0.998 / Rmerge(I) obs: 0.116 / Net I/σ(I): 6.5 |
| Reflection shell | Resolution: 3.2→3.37 Å / Rmerge(I) obs: 0.626 / Mean I/σ(I) obs: 1.8 / Num. unique obs: 3109 / CC1/2: 0.526 / % possible all: 96.7 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5NX2 Resolution: 3.2→30 Å / Cor.coef. Fo:Fc: 0.924 / Cor.coef. Fo:Fc free: 0.886 / Rfactor Rfree error: 0 / Cross valid method: THROUGHOUT / σ(F): 0 / SU Rfree Blow DPI: 0.453
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 261 Å2 / Biso mean: 136.81 Å2 / Biso min: 58.31 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3.2→30 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.2→3.36 Å / Rfactor Rfree error: 0 / Total num. of bins used: 11
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj








