[English] 日本語
 Yorodumi
Yorodumi- PDB-6k4z: Single-chain Fv antibody of C6 COMPLEXED WITH 2-(4-HYDROXY-3-NITR... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6k4z | ||||||
|---|---|---|---|---|---|---|---|
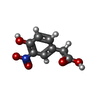
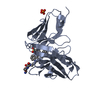
| Title | Single-chain Fv antibody of C6 COMPLEXED WITH 2-(4-HYDROXY-3-NITROPHENYL)ACETIC ACID | ||||||
 Components Components | scFv of C6 | ||||||
 Keywords Keywords |  IMMUNE SYSTEM / IMMUNE SYSTEM /  antigen binding / antigen binding /  affinity maturation / affinity maturation /  Somatic hypermutation / Somatic hypermutation /  conformational change conformational change | ||||||
| Function / homology | 2-(4-HYDROXY-3-NITROPHENYL)ACETIC ACID Function and homology information Function and homology information | ||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.65 Å MOLECULAR REPLACEMENT / Resolution: 1.65 Å | ||||||
 Authors Authors | Nishiguchi, A. / Numoto, N. / Ito, N. / Azuma, T. / Oda, M. | ||||||
 Citation Citation |  Journal: Mol.Immunol. / Year: 2019 Journal: Mol.Immunol. / Year: 2019Title: Three-dimensional structure of a high affinity anti-(4-hydroxy-3-nitrophenyl)acetyl antibody possessing a glycine residue at position 95 of the heavy chain. Authors: Nishiguchi, A. / Numoto, N. / Ito, N. / Azuma, T. / Oda, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6k4z.cif.gz 6k4z.cif.gz | 118.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6k4z.ent.gz pdb6k4z.ent.gz | 87 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6k4z.json.gz 6k4z.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k4/6k4z https://data.pdbj.org/pub/pdb/validation_reports/k4/6k4z ftp://data.pdbj.org/pub/pdb/validation_reports/k4/6k4z ftp://data.pdbj.org/pub/pdb/validation_reports/k4/6k4z | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3et9S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
|
- Components
Components
| #1: Antibody | Mass: 26116.920 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: The fusion protein of expression tags (residues -2-0), the light chain (residues 1-108), linker GGGGSGGGGSGGGGS (residues 501-515), and the heavy chain (residues 1001-1113) Source: (gene. exp.)   Mus musculus (house mouse) / Plasmid: pET-32TH / Production host: Mus musculus (house mouse) / Plasmid: pET-32TH / Production host:   Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3) / Variant (production host): codon plus Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3) / Variant (production host): codon plus#2: Chemical | #3: Chemical | ChemComp-SO4 / |  Sulfate Sulfate#4: Chemical | ChemComp-GOL / |  Glycerol Glycerol#5: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.37 Å3/Da / Density % sol: 48.13 % |
|---|---|
Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 8.5 Details: 0.14 M Lithium sulfate monohydrate, 0.07 M TRIS hydrochloride pH 8.5, 21% w/v Polyethylene glycol 4000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Photon Factory Photon Factory  / Beamline: BL-1A / Wavelength: 1.1 Å / Beamline: BL-1A / Wavelength: 1.1 Å |
| Detector | Type: DECTRIS EIGER X 4M / Detector: PIXEL / Date: Dec 12, 2018 |
| Radiation | Monochromator: Cryo-cooled channel-cut Si(111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.1 Å / Relative weight: 1 : 1.1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.65→50 Å / Num. obs: 52628 / % possible obs: 96.2 % / Redundancy: 3.6 % / Biso Wilson estimate: 17.07 Å2 / CC1/2: 0.998 / Rsym value: 0.073 / Net I/σ(I): 11.2 |
| Reflection shell | Resolution: 1.65→1.75 Å / Redundancy: 3.5 % / Num. unique obs: 8339 / CC1/2: 0.716 / Rsym value: 0.586 / % possible all: 94.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3ET9 Resolution: 1.65→43.29 Å / SU ML: 0.173 / Cross valid method: FREE R-VALUE / σ(F): 1.96 / Phase error: 21.1576
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 22.58 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.65→43.29 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj