[English] 日本語
 Yorodumi
Yorodumi- PDB-6j0k: Crystal structure of intracellular B30.2 domain of BTN3A3 mutant ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6j0k | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with HMBPP | ||||||
 Components Components | Butyrophilin subfamily 3 member A3 | ||||||
 Keywords Keywords |  SIGNALING PROTEIN / SIGNALING PROTEIN /  Butyrophilin Butyrophilin | ||||||
| Function / homology |  Function and homology information Function and homology informationButyrophilin (BTN) family interactions / T cell mediated immunity / regulation of cytokine production / T cell receptor signaling pathway / external side of plasma membrane /  signaling receptor binding / signaling receptor binding /  membrane / membrane /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2 Å MOLECULAR REPLACEMENT / Resolution: 2 Å | ||||||
 Authors Authors | Yang, Y.Y. / Liu, W.D. / Cai, N.N. / Chen, C.C. / Guo, R.T. / Zhang, Y.H. | ||||||
 Citation Citation |  Journal: Immunity / Year: 2019 Journal: Immunity / Year: 2019Title: A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation. Authors: Yang, Y. / Li, L. / Yuan, L. / Zhou, X. / Duan, J. / Xiao, H. / Cai, N. / Han, S. / Ma, X. / Liu, W. / Chen, C.C. / Wang, L. / Li, X. / Chen, J. / Kang, N. / Chen, J. / Shen, Z. / Malwal, S. ...Authors: Yang, Y. / Li, L. / Yuan, L. / Zhou, X. / Duan, J. / Xiao, H. / Cai, N. / Han, S. / Ma, X. / Liu, W. / Chen, C.C. / Wang, L. / Li, X. / Chen, J. / Kang, N. / Chen, J. / Shen, Z. / Malwal, S.R. / Liu, W. / Shi, Y. / Oldfield, E. / Guo, R.T. / Zhang, Y. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6j0k.cif.gz 6j0k.cif.gz | 97.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6j0k.ent.gz pdb6j0k.ent.gz | 71.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6j0k.json.gz 6j0k.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/j0/6j0k https://data.pdbj.org/pub/pdb/validation_reports/j0/6j0k ftp://data.pdbj.org/pub/pdb/validation_reports/j0/6j0k ftp://data.pdbj.org/pub/pdb/validation_reports/j0/6j0k | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5zxkC  5zz3C  6ismC  6itaC  6j06C  6j0gC  6j0lC  4n7uS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
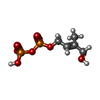
| #1: Protein | Mass: 25272.418 Da / Num. of mol.: 2 / Fragment: UNP residues 328-515 / Mutation: R351H Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: BTN3A3, BTF3 / Production host: Homo sapiens (human) / Gene: BTN3A3, BTF3 / Production host:   Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3) / References: UniProt: O00478 Escherichia coli BL21(DE3) (bacteria) / Strain (production host): BL21(DE3) / References: UniProt: O00478#2: Chemical |  (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate#3: Water | ChemComp-HOH / |  Water WaterSequence details | The intracellular B30.2 domain of BTN3A3 was redesigned to have TEV protease cleavage sites and ...The intracellular B30.2 domain of BTN3A3 was redesigned to have TEV protease cleavage sites and bordering the linker region(AGAGA). | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.76 Å3/Da / Density % sol: 30.22 % |
|---|---|
Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / Details: Isopropanol ,Imidazole, PEG 8000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSRRC NSRRC  / Beamline: BL15A1 / Wavelength: 1 Å / Beamline: BL15A1 / Wavelength: 1 Å |
| Detector | Type: RAYONIX MX-300 / Detector: CCD / Date: Nov 29, 2017 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2→25 Å / Num. obs: 20422 / % possible obs: 84.2 % / Redundancy: 3 % / Rmerge(I) obs: 0.08 / Net I/σ(I): 16.06 |
| Reflection shell | Resolution: 2→2.07 Å / Num. unique obs: 2264 / CC1/2: 0.946 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 4N7U Resolution: 2→25 Å / Cor.coef. Fo:Fc: 0.954 / Cor.coef. Fo:Fc free: 0.912 / SU B: 4.815 / SU ML: 0.139 / Cross valid method: THROUGHOUT / ESU R: 0.299 / ESU R Free: 0.226 / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 25.298 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 2→25 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj