[English] 日本語
 Yorodumi
Yorodumi- PDB-6ir7: Green fluorescent protein variant GFPuv with the modification to ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ir7 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Green fluorescent protein variant GFPuv with the modification to 6-hydroxynorleucine at the C-terminus | |||||||||
 Components Components | Green fluorescent protein | |||||||||
 Keywords Keywords |  FLUORESCENT PROTEIN FLUORESCENT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Aequorea victoria (jellyfish) Aequorea victoria (jellyfish) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.277 Å MOLECULAR REPLACEMENT / Resolution: 1.277 Å | |||||||||
 Authors Authors | Nakatani, T. / Yasui, N. / Yamashita, A. | |||||||||
| Funding support |  Japan, 2items Japan, 2items
| |||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2019 Journal: Sci Rep / Year: 2019Title: Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli. Authors: Nakatani, T. / Yasui, N. / Tamura, I. / Yamashita, A. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ir7.cif.gz 6ir7.cif.gz | 118.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ir7.ent.gz pdb6ir7.ent.gz | 88.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ir7.json.gz 6ir7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ir/6ir7 https://data.pdbj.org/pub/pdb/validation_reports/ir/6ir7 ftp://data.pdbj.org/pub/pdb/validation_reports/ir/6ir7 ftp://data.pdbj.org/pub/pdb/validation_reports/ir/6ir7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6ir6C  1b9cS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  Mass: 26681.920 Da / Num. of mol.: 1 / Mutation: Q80R, F99S, M153T, V163A, A206K Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Aequorea victoria (jellyfish) / Gene: GFP / Plasmid: pET25b / Production host: Aequorea victoria (jellyfish) / Gene: GFP / Plasmid: pET25b / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21 (DE3) pLysS / References: UniProt: P42212 Escherichia coli (E. coli) / Strain (production host): BL21 (DE3) pLysS / References: UniProt: P42212 |
|---|---|
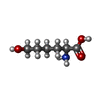
| #2: Chemical | ChemComp-LDO / |
| #3: Chemical | ChemComp-SO4 /  Sulfate Sulfate |
| #4: Chemical | ChemComp-MES /  MES (buffer) MES (buffer) |
| #5: Water | ChemComp-HOH /  Water Water |
| Sequence details | RESIDUE THR 65 HAS BEEN MUTATED TO SER 65. RESIDUES SER 65, TYR 66 AND GLY 67 CONSTITUTE THE ...RESIDUE THR 65 HAS BEEN MUTATED TO SER 65. RESIDUES SER 65, TYR 66 AND GLY 67 CONSTITUTE |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.15 Å3/Da / Density % sol: 42.78 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop Details: 0.2 M Ammonium Sulfate, 0.1 M MES pH 6.5, 30% PEGMME5000 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SPring-8 SPring-8  / Beamline: BL41XU / Wavelength: 1 Å / Beamline: BL41XU / Wavelength: 1 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: Jul 18, 2014 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 1.28→50 Å / Num. obs: 56911 / % possible obs: 96.3 % / Redundancy: 3.1 % / Rsym value: 0.071 / Net I/σ(I): 27.8 |
| Reflection shell | Resolution: 1.28→1.3 Å / Redundancy: 2.9 % / Mean I/σ(I) obs: 7.2 / Num. unique obs: 2566 / Rsym value: 0.248 / % possible all: 87.4 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1B9C Resolution: 1.277→47.649 Å / SU ML: 0.11 / Cross valid method: FREE R-VALUE / σ(F): 1.4 / Phase error: 18.24 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.277→47.649 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj