+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6gh9 | ||||||
|---|---|---|---|---|---|---|---|
| Title | USP15 catalytic domain in complex with small molecule | ||||||
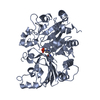
 Components Components | Ubiquitin carboxyl-terminal hydrolase 15 | ||||||
 Keywords Keywords |  HYDROLASE / HYDROLASE /  protease / protease /  ubiquitin / ubiquitin specific protease / ubiquitin / ubiquitin specific protease /  inhibitor / inhibitor /  mitoxantrone mitoxantrone | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator / negative regulation of antifungal innate immune response / protein K27-linked deubiquitination / positive regulation of RIG-I signaling pathway / ubiquitin-modified histone reader activity / monoubiquitinated protein deubiquitination /  deubiquitinase activity / deubiquitinase activity /  transforming growth factor beta receptor binding / K48-linked deubiquitinase activity / transcription elongation-coupled chromatin remodeling ...regulation of intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator / negative regulation of antifungal innate immune response / protein K27-linked deubiquitination / positive regulation of RIG-I signaling pathway / ubiquitin-modified histone reader activity / monoubiquitinated protein deubiquitination / transforming growth factor beta receptor binding / K48-linked deubiquitinase activity / transcription elongation-coupled chromatin remodeling ...regulation of intrinsic apoptotic signaling pathway in response to osmotic stress by p53 class mediator / negative regulation of antifungal innate immune response / protein K27-linked deubiquitination / positive regulation of RIG-I signaling pathway / ubiquitin-modified histone reader activity / monoubiquitinated protein deubiquitination /  deubiquitinase activity / deubiquitinase activity /  transforming growth factor beta receptor binding / K48-linked deubiquitinase activity / transcription elongation-coupled chromatin remodeling / protein deubiquitination / SMAD binding / BMP signaling pathway / Downregulation of TGF-beta receptor signaling / transforming growth factor beta receptor signaling pathway / negative regulation of transforming growth factor beta receptor signaling pathway / UCH proteinases / transforming growth factor beta receptor binding / K48-linked deubiquitinase activity / transcription elongation-coupled chromatin remodeling / protein deubiquitination / SMAD binding / BMP signaling pathway / Downregulation of TGF-beta receptor signaling / transforming growth factor beta receptor signaling pathway / negative regulation of transforming growth factor beta receptor signaling pathway / UCH proteinases /  ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity /  nuclear body / Ub-specific processing proteases / cysteine-type endopeptidase activity / nuclear body / Ub-specific processing proteases / cysteine-type endopeptidase activity /  mitochondrion / mitochondrion /  proteolysis / proteolysis /  nucleoplasm / identical protein binding / nucleoplasm / identical protein binding /  nucleus / nucleus /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.09 Å MOLECULAR REPLACEMENT / Resolution: 2.09 Å | ||||||
 Authors Authors | Ward, S.J. / Gratton, H.E. / Caulton, S.G. / Emsley, J. / Dreveny, I. | ||||||
| Funding support |  United Kingdom, 1items United Kingdom, 1items
| ||||||
 Citation Citation |  Journal: J. Biol. Chem. / Year: 2018 Journal: J. Biol. Chem. / Year: 2018Title: The structure of the deubiquitinase USP15 reveals a misaligned catalytic triad and an open ubiquitin-binding channel. Authors: Ward, S.J. / Gratton, H.E. / Indrayudha, P. / Michavila, C. / Mukhopadhyay, R. / Maurer, S.K. / Caulton, S.G. / Emsley, J. / Dreveny, I. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6gh9.cif.gz 6gh9.cif.gz | 278 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6gh9.ent.gz pdb6gh9.ent.gz | 223.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6gh9.json.gz 6gh9.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/gh/6gh9 https://data.pdbj.org/pub/pdb/validation_reports/gh/6gh9 ftp://data.pdbj.org/pub/pdb/validation_reports/gh/6gh9 ftp://data.pdbj.org/pub/pdb/validation_reports/gh/6gh9 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6ghaC  2y6eS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Ens-ID: 1
|
 Movie
Movie Controller
Controller












 PDBj
PDBj


