[English] 日本語
 Yorodumi
Yorodumi- PDB-6del: Crystal structure of Candida albicans acetohydroxyacid synthase i... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6del | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Candida albicans acetohydroxyacid synthase in complex with the herbicide chlorimuron ethyl | ||||||
 Components Components | Acetolactate synthase | ||||||
 Keywords Keywords | transferase/transferase inhibitor / AHAS /  acetohydroxyacid synthase / acetohydroxyacid synthase /  acetolactate synthase / acetolactate synthase /  herbicide / herbicide /  thiamin diphosphate / FAD / thiamin diphosphate / FAD /  transferase / chlorimuron ethyl / transferase / chlorimuron ethyl /  sulfonylurea / transferase-transferase inhibitor complex sulfonylurea / transferase-transferase inhibitor complex | ||||||
| Function / homology |  Function and homology information Function and homology information acetolactate synthase / acetolactate synthase /  acetolactate synthase complex / acetolactate synthase complex /  acetolactate synthase activity / valine biosynthetic process / isoleucine biosynthetic process / acetolactate synthase activity / valine biosynthetic process / isoleucine biosynthetic process /  thiamine pyrophosphate binding / amino acid biosynthetic process / thiamine pyrophosphate binding / amino acid biosynthetic process /  flavin adenine dinucleotide binding / magnesium ion binding / flavin adenine dinucleotide binding / magnesium ion binding /  mitochondrion mitochondrionSimilarity search - Function | ||||||
| Biological species |   Candida albicans (yeast) Candida albicans (yeast) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.119 Å MOLECULAR REPLACEMENT / Resolution: 2.119 Å | ||||||
 Authors Authors | Garcia, M.D. / Guddat, L.W. | ||||||
| Funding support |  Australia, 1items Australia, 1items
| ||||||
 Citation Citation |  Journal: Proc. Natl. Acad. Sci. U.S.A. / Year: 2018 Journal: Proc. Natl. Acad. Sci. U.S.A. / Year: 2018Title: Commercial AHAS-inhibiting herbicides are promising drug leads for the treatment of human fungal pathogenic infections. Authors: Garcia, M.D. / Chua, S.M.H. / Low, Y.S. / Lee, Y.T. / Agnew-Francis, K. / Wang, J.G. / Nouwens, A. / Lonhienne, T. / Williams, C.M. / Fraser, J.A. / Guddat, L.W. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6del.cif.gz 6del.cif.gz | 153.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6del.ent.gz pdb6del.ent.gz | 113.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6del.json.gz 6del.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/de/6del https://data.pdbj.org/pub/pdb/validation_reports/de/6del ftp://data.pdbj.org/pub/pdb/validation_reports/de/6del ftp://data.pdbj.org/pub/pdb/validation_reports/de/6del | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6dekSC  6demC  6denC  6deoC  6depC  6deqC  6derC  6desC S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein |  Mass: 74315.367 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Candida albicans (strain SC5314 / ATCC MYA-2876) (yeast) Candida albicans (strain SC5314 / ATCC MYA-2876) (yeast)Strain: SC5314 / ATCC MYA-2876 / Gene: ILV2, orf19.1613, CAALFM_C302320WA / Plasmid: pET30a(+) / Production host:   Escherichia coli BL21(DE3) (bacteria) / References: UniProt: A0A1D8PJF9, Escherichia coli BL21(DE3) (bacteria) / References: UniProt: A0A1D8PJF9,  acetolactate synthase acetolactate synthase |
|---|
-Non-polymers , 8 types, 563 molecules 



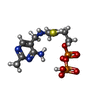










| #2: Chemical | ChemComp-K / |
|---|---|
| #3: Chemical | ChemComp-MG / |
| #4: Chemical | ChemComp-CIE / |
| #5: Chemical | ChemComp-FAD /  Flavin adenine dinucleotide Flavin adenine dinucleotide |
| #6: Chemical | ChemComp-TP9 / ( |
| #7: Chemical | ChemComp-G8G / ( |
| #8: Chemical | ChemComp-FMT /  Formic acid Formic acid |
| #9: Water | ChemComp-HOH /  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 5.37 Å3/Da / Density % sol: 77 % |
|---|---|
Crystal grow | Temperature: 290 K / Method: vapor diffusion, hanging drop Details: 1 mM FAD, 1 mM ThDP, 5 mM MgCl2, 5 mM DTT, 3 mM sodium pyruvate, 1 M Na/K tartrate, 0.1 M CHES, and 0.2 M ammonium sulfate, 1 mM chlorimuron ethyl PH range: 9.4-9.8 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Australian Synchrotron Australian Synchrotron  / Beamline: MX1 / Wavelength: 0.9537 Å / Beamline: MX1 / Wavelength: 0.9537 Å |
| Detector | Type: ADSC QUANTUM 210r / Detector: CCD / Date: Mar 19, 2016 / Details: Mirrors |
| Radiation | Monochromator: SI(III) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9537 Å / Relative weight: 1 : 0.9537 Å / Relative weight: 1 |
| Reflection | Resolution: 2.119→48.39 Å / Num. obs: 91976 / % possible obs: 99.7 % / Redundancy: 21.4 % / Rmerge(I) obs: 0.161 / Rpim(I) all: 0.051 / Rrim(I) all: 0.169 / Net I/σ(I): 11.3 |
| Reflection shell | Resolution: 2.12→2.16 Å / Redundancy: 18.7 % / Rmerge(I) obs: 0.895 / Num. unique obs: 4219 / Rpim(I) all: 0.291 / Rrim(I) all: 0.942 / % possible all: 93.7 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6DEK Resolution: 2.119→46.794 Å / SU ML: 0.2 / Cross valid method: FREE R-VALUE / σ(F): 1.33 / Phase error: 20.21 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.119→46.794 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj










