+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6bcs | ||||||
|---|---|---|---|---|---|---|---|
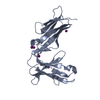
| Title | LilrB2 D1D2 domains complexed with benzamidine | ||||||
 Components Components | Leukocyte immunoglobulin-like receptor subfamily B member 2 | ||||||
 Keywords Keywords |  IMMUNE SYSTEM / IMMUNE SYSTEM /  receptor / IG LIKE DOMAINS receptor / IG LIKE DOMAINS | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of antigen processing and presentation / negative regulation of postsynaptic density organization / interleukin-10-mediated signaling pathway / negative regulation of T cell costimulation / positive regulation of tolerance induction / MHC class Ib protein complex binding / inhibitory MHC class I receptor activity / immune response-inhibiting cell surface receptor signaling pathway / MHC class Ib protein binding / Fc receptor mediated inhibitory signaling pathway ...negative regulation of antigen processing and presentation / negative regulation of postsynaptic density organization / interleukin-10-mediated signaling pathway / negative regulation of T cell costimulation / positive regulation of tolerance induction / MHC class Ib protein complex binding / inhibitory MHC class I receptor activity / immune response-inhibiting cell surface receptor signaling pathway / MHC class Ib protein binding / Fc receptor mediated inhibitory signaling pathway / positive regulation of long-term synaptic depression / positive regulation of T cell tolerance induction /  protein phosphatase 1 binding / positive regulation of regulatory T cell differentiation / regulation of dendritic cell differentiation / regulation of long-term synaptic potentiation / heterotypic cell-cell adhesion / negative regulation of protein metabolic process / MHC class I protein binding / tertiary granule membrane / negative regulation of calcium ion transport / ficolin-1-rich granule membrane / cellular defense response / positive regulation of T cell proliferation / negative regulation of T cell proliferation / positive regulation of protein dephosphorylation / protein phosphatase 1 binding / positive regulation of regulatory T cell differentiation / regulation of dendritic cell differentiation / regulation of long-term synaptic potentiation / heterotypic cell-cell adhesion / negative regulation of protein metabolic process / MHC class I protein binding / tertiary granule membrane / negative regulation of calcium ion transport / ficolin-1-rich granule membrane / cellular defense response / positive regulation of T cell proliferation / negative regulation of T cell proliferation / positive regulation of protein dephosphorylation /  cell adhesion molecule binding / cytokine-mediated signaling pathway / positive regulation of interleukin-6 production / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / cell-cell signaling / cell adhesion molecule binding / cytokine-mediated signaling pathway / positive regulation of interleukin-6 production / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / cell-cell signaling /  amyloid-beta binding / amyloid-beta binding /  adaptive immune response / cellular response to lipopolysaccharide / membrane => GO:0016020 / learning or memory / cell surface receptor signaling pathway / adaptive immune response / cellular response to lipopolysaccharide / membrane => GO:0016020 / learning or memory / cell surface receptor signaling pathway /  immune response / Neutrophil degranulation / protein-containing complex binding / immune response / Neutrophil degranulation / protein-containing complex binding /  cell surface / cell surface /  signal transduction / protein homodimerization activity / signal transduction / protein homodimerization activity /  extracellular space / extracellular space /  membrane / membrane /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.1 Å molecular replacement / Resolution: 2.1 Å | ||||||
 Authors Authors | Cao, Q. / Sawaya, M.R. / Eisenberg, D.S. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Nat Chem / Year: 2018 Journal: Nat Chem / Year: 2018Title: Inhibiting amyloid-beta cytotoxicity through its interaction with the cell surface receptor LilrB2 by structure-based design. Authors: Cao, Q. / Shin, W.S. / Chan, H. / Vuong, C.K. / Dubois, B. / Li, B. / Murray, K.A. / Sawaya, M.R. / Feigon, J. / Black, D.L. / Eisenberg, D.S. / Jiang, L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6bcs.cif.gz 6bcs.cif.gz | 95.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6bcs.ent.gz pdb6bcs.ent.gz | 69.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6bcs.json.gz 6bcs.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bc/6bcs https://data.pdbj.org/pub/pdb/validation_reports/bc/6bcs ftp://data.pdbj.org/pub/pdb/validation_reports/bc/6bcs ftp://data.pdbj.org/pub/pdb/validation_reports/bc/6bcs | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  2gw5S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 22065.891 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: LILRB2 / Plasmid: pET28a / Production host: Homo sapiens (human) / Gene: LILRB2 / Plasmid: pET28a / Production host:   Escherichia coli BL21(DE3) (bacteria) / References: UniProt: A0A0G2JNQ7, UniProt: Q8N423*PLUS Escherichia coli BL21(DE3) (bacteria) / References: UniProt: A0A0G2JNQ7, UniProt: Q8N423*PLUS | ||||||
|---|---|---|---|---|---|---|---|
| #2: Chemical | ChemComp-BEN /  Benzamidine Benzamidine#3: Chemical |  Chloride Chloride#4: Chemical |  Dimethyl sulfoxide Dimethyl sulfoxide#5: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.87 Å3/Da / Density % sol: 34.07 % |
|---|---|
Crystal grow | Temperature: 298 K / Method: vapor diffusion, hanging drop / pH: 5.53 Details: 1.03 M Ammonium sulfate, 0.1 M Bis-Tris pH 5.53, 2.76% PEG 3350, 1.4% w/v benzamidine |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 24-ID-C / Wavelength: 0.9795 Å / Beamline: 24-ID-C / Wavelength: 0.9795 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS 6M-F / Detector: PIXEL / Date: Oct 22, 2016 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 0.9795 Å / Relative weight: 1 : 0.9795 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.1→49.695 Å / Num. obs: 10310 / % possible obs: 99.9 % / Observed criterion σ(I): -3 / Redundancy: 12.269 % / Biso Wilson estimate: 34.75 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.089 / Rrim(I) all: 0.093 / Χ2: 0.896 / Net I/σ(I): 17.98 / Num. measured all: 126498 / Scaling rejects: 55 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR | Model details: Phaser MODE: MR_AUTO
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 2GW5 Resolution: 2.1→49.695 Å / SU ML: 0.25 / Cross valid method: FREE R-VALUE / σ(F): 1.36 / Phase error: 24.35
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 97.14 Å2 / Biso mean: 37.5477 Å2 / Biso min: 17.34 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.1→49.695 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Rfactor Rfree error: 0 / Total num. of bins used: 7 / % reflection obs: 100 %
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: -24.2388 Å / Origin y: 10.7851 Å / Origin z: -2.4184 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj












