+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5xls | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of UraA in occluded conformation | |||||||||
 Components Components | Uracil permease | |||||||||
 Keywords Keywords |  TRANSPORT PROTEIN / Occluded conformation / dimer TRANSPORT PROTEIN / Occluded conformation / dimer | |||||||||
| Function / homology |  Function and homology information Function and homology informationuracil transmembrane transporter activity / membrane => GO:0016020 /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Escherichia coli O157:H7 (bacteria) Escherichia coli O157:H7 (bacteria) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.5 Å MOLECULAR REPLACEMENT / Resolution: 2.5 Å | |||||||||
 Authors Authors | Yu, X.Z. / Yang, G.H. / Yan, C.Y. / Yan, N. | |||||||||
| Funding support |  China, 2items China, 2items
| |||||||||
 Citation Citation |  Journal: Cell Res. / Year: 2017 Journal: Cell Res. / Year: 2017Title: Dimeric structure of the uracil:proton symporter UraA provides mechanistic insights into the SLC4/23/26 transporters Authors: Yu, X. / Yang, G. / Yan, C. / Baylon, J.L. / Jiang, J. / Fan, H. / Lu, G. / Hasegawa, K. / Okumura, H. / Wang, T. / Tajkhorshid, E. / Li, S. / Yan, N. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5xls.cif.gz 5xls.cif.gz | 172.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5xls.ent.gz pdb5xls.ent.gz | 137.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5xls.json.gz 5xls.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/xl/5xls https://data.pdbj.org/pub/pdb/validation_reports/xl/5xls ftp://data.pdbj.org/pub/pdb/validation_reports/xl/5xls ftp://data.pdbj.org/pub/pdb/validation_reports/xl/5xls | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3qe7S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 45946.723 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Escherichia coli O157:H7 (bacteria) / Gene: uraA, Z3760, ECs3359 / Production host: Escherichia coli O157:H7 (bacteria) / Gene: uraA, Z3760, ECs3359 / Production host:   Escherichia coli (E. coli) / References: UniProt: P0AGM8 Escherichia coli (E. coli) / References: UniProt: P0AGM8 | ||
|---|---|---|---|
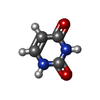
| #2: Chemical | ChemComp-URA /  Uracil Uracil | ||
| #3: Chemical | | #4: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.93 Å3/Da / Density % sol: 68.69 % |
|---|---|
Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 24% PEG 400, 100mM MES-NaOH, pH 6.5, 100mM NaF, 50mM MgCl2, 3mM (NH4)2WS4 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SPring-8 SPring-8  / Beamline: BL41XU / Wavelength: 1 Å / Beamline: BL41XU / Wavelength: 1 Å |
| Detector | Type: RAYONIX MX225HE / Detector: CCD / Date: Oct 5, 2012 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.5→40 Å / Num. obs: 49846 / % possible obs: 98 % / Redundancy: 3.3 % / Rsym value: 0.117 / Net I/σ(I): 18.5 |
| Reflection shell | Resolution: 2.5→2.59 Å / Redundancy: 3.3 % / Rmerge(I) obs: 0.558 / Mean I/σ(I) obs: 2.8 / % possible all: 98.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3QE7 Resolution: 2.5→32.79 Å / Cross valid method: FREE R-VALUE / σ(F): 0.88 Details: THE STRUCTURE FACTOR FILE CONTAINS FRIEDEL PAIRS IN F_PLUS/MINUS AND I_PLUS/MINUS COLUMNS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.47 Å / VDW probe radii: 0.8 Å / Bsol: 42.511 Å2 / ksol: 0.313 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.5→32.79 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Origin x: 19.2014 Å / Origin y: 15.5427 Å / Origin z: -12.0268 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group | Selection details: all |
 Movie
Movie Controller
Controller













 PDBj
PDBj