| Entry | Database: PDB / ID: 5vqb
|
|---|
| Title | Crystal structure of rifampin monooxygenase from Streptomyces venezuelae, complex with FAD |
|---|
 Components Components | Rifampin monooxygenase |
|---|
 Keywords Keywords |  OXIDOREDUCTASE / OXIDOREDUCTASE /  rifampin / rifampin /  rifamycin / rifamycin /  antibiotic resistance / antibiotic resistance /  monooxygenase / FAD / monooxygenase / FAD /  flavin adenine dinucleotide / flavin adenine dinucleotide /  structural genomics / CSGID / Center for Structural Genomics of Infectious Diseases / NIAID / structural genomics / CSGID / Center for Structural Genomics of Infectious Diseases / NIAID /  National Institute of Allergy and Infectious Disease National Institute of Allergy and Infectious Disease |
|---|
| Function / homology |  Function and homology information Function and homology information
Alpha-Beta Plaits - #2450 / Glutaredoxin - #120 / FAD-binding domain / FAD binding domain / FAD/NAD(P)-binding domain / FAD/NAD(P)-binding domain / 3-Layer(bba) Sandwich / FAD/NAD(P)-binding domain superfamily /  Glutaredoxin / Alpha-Beta Plaits ...Alpha-Beta Plaits - #2450 / Glutaredoxin - #120 / FAD-binding domain / FAD binding domain / FAD/NAD(P)-binding domain / FAD/NAD(P)-binding domain / 3-Layer(bba) Sandwich / FAD/NAD(P)-binding domain superfamily / Glutaredoxin / Alpha-Beta Plaits ...Alpha-Beta Plaits - #2450 / Glutaredoxin - #120 / FAD-binding domain / FAD binding domain / FAD/NAD(P)-binding domain / FAD/NAD(P)-binding domain / 3-Layer(bba) Sandwich / FAD/NAD(P)-binding domain superfamily /  Glutaredoxin / Alpha-Beta Plaits / 2-Layer Sandwich / 3-Layer(aba) Sandwich / Alpha BetaSimilarity search - Domain/homology Glutaredoxin / Alpha-Beta Plaits / 2-Layer Sandwich / 3-Layer(aba) Sandwich / Alpha BetaSimilarity search - Domain/homology |
|---|
| Biological species |   Streptomyces venezuelae (bacteria) Streptomyces venezuelae (bacteria) |
|---|
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 3.391 Å MOLECULAR REPLACEMENT / Resolution: 3.391 Å |
|---|
 Authors Authors | Cox, G. / Kelso, J. / Stogios, P.J. / Savchenko, A. / Anderson, W.F. / Wright, G.D. / Center for Structural Genomics of Infectious Diseases (CSGID) |
|---|
| Funding support |  United States, 1items United States, 1items | Organization | Grant number | Country |
|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | HHSN27220120026C |  United States United States |
|
|---|
 Citation Citation |  Journal: Cell Chem Biol / Year: 2018 Journal: Cell Chem Biol / Year: 2018
Title: Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action.
Authors: Koteva, K. / Cox, G. / Kelso, J.K. / Surette, M.D. / Zubyk, H.L. / Ejim, L. / Stogios, P. / Savchenko, A. / Sorensen, D. / Wright, G.D. |
|---|
| History | | Deposition | May 8, 2017 | Deposition site: RCSB / Processing site: RCSB |
|---|
| Revision 1.0 | Aug 16, 2017 | Provider: repository / Type: Initial release |
|---|
| Revision 1.1 | Sep 13, 2017 | Group: Author supporting evidence / Category: pdbx_audit_support / Item: _pdbx_audit_support.funding_organization |
|---|
| Revision 1.2 | Feb 21, 2018 | Group: Database references
Category: citation / citation_author / pdbx_database_related
Item: _citation.country / _citation.journal_abbrev ..._citation.country / _citation.journal_abbrev / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _pdbx_database_related.db_id |
|---|
| Revision 1.3 | May 2, 2018 | Group: Data collection / Database references / Category: citation
Item: _citation.journal_volume / _citation.page_first / _citation.page_last |
|---|
| Revision 1.4 | Dec 11, 2019 | Group: Author supporting evidence / Category: pdbx_audit_support / Item: _pdbx_audit_support.funding_organization |
|---|
| Revision 1.5 | Oct 4, 2023 | Group: Data collection / Database references / Refinement description
Category: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords OXIDOREDUCTASE /
OXIDOREDUCTASE /  rifampin /
rifampin /  rifamycin /
rifamycin /  antibiotic resistance /
antibiotic resistance /  monooxygenase / FAD /
monooxygenase / FAD /  flavin adenine dinucleotide /
flavin adenine dinucleotide /  structural genomics / CSGID / Center for Structural Genomics of Infectious Diseases / NIAID /
structural genomics / CSGID / Center for Structural Genomics of Infectious Diseases / NIAID /  National Institute of Allergy and Infectious Disease
National Institute of Allergy and Infectious Disease Function and homology information
Function and homology information
 Streptomyces venezuelae (bacteria)
Streptomyces venezuelae (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 3.391 Å
MOLECULAR REPLACEMENT / Resolution: 3.391 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: Cell Chem Biol / Year: 2018
Journal: Cell Chem Biol / Year: 2018 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5vqb.cif.gz
5vqb.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5vqb.ent.gz
pdb5vqb.ent.gz PDB format
PDB format 5vqb.json.gz
5vqb.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/vq/5vqb
https://data.pdbj.org/pub/pdb/validation_reports/vq/5vqb ftp://data.pdbj.org/pub/pdb/validation_reports/vq/5vqb
ftp://data.pdbj.org/pub/pdb/validation_reports/vq/5vqb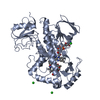

 Links
Links Assembly
Assembly



 Components
Components
 Streptomyces venezuelae (strain ATCC 10712 / CBS 650.69 / DSM 40230 / JCM 4526 / NBRC 13096 / PD 04745) (bacteria)
Streptomyces venezuelae (strain ATCC 10712 / CBS 650.69 / DSM 40230 / JCM 4526 / NBRC 13096 / PD 04745) (bacteria)
 Escherichia coli (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: F2R776
Escherichia coli (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: F2R776 Flavin adenine dinucleotide
Flavin adenine dinucleotide Chloride
Chloride Glycerol
Glycerol Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å
ROTATING ANODE / Type: RIGAKU MICROMAX-007 HF / Wavelength: 1.5418 Å : 1.5418 Å / Relative weight: 1
: 1.5418 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj





