[English] 日本語
 Yorodumi
Yorodumi- PDB-5tcm: First Bromodomain from Leishmania donovani LdBPK.091320 complexed... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5tcm | ||||||
|---|---|---|---|---|---|---|---|
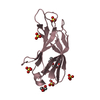
| Title | First Bromodomain from Leishmania donovani LdBPK.091320 complexed with BI-2536 | ||||||
 Components Components | Uncharacterized protein | ||||||
 Keywords Keywords | UNKNOWN FUNCTION /  Bromodomain / Inhibitor / Structural Genomics Consortium (SGC) Bromodomain / Inhibitor / Structural Genomics Consortium (SGC) | ||||||
| Function / homology |  Function and homology information Function and homology informationlysine-acetylated histone binding / regulation of transcription by RNA polymerase II /  nucleus nucleusSimilarity search - Function | ||||||
| Biological species |   Leishmania donovani (eukaryote) Leishmania donovani (eukaryote) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.2 Å molecular replacement / Resolution: 2.2 Å | ||||||
 Authors Authors | Lin, Y.H. / Hou, C.F.D. / Loppnau, P. / Dong, A. / Bountra, C. / Edwards, A.M. / Arrowsmith, C.H. / Hui, R. / Walker, J.R. / Structural Genomics Consortium (SGC) | ||||||
 Citation Citation |  Journal: To be published Journal: To be publishedTitle: First Bromodomain from Leishmania donovani LdBPK.091320 complexed with BI-2536 Authors: Lin, Y.H. / Hou, C.F.D. / Loppnau, P. / Dong, A. / Bountra, C. / Edwards, A.M. / Arrowsmith, C.H. / Hui, R. / Walker, J.R. / Structural Genomics Consortium (SGC) | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5tcm.cif.gz 5tcm.cif.gz | 157.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5tcm.ent.gz pdb5tcm.ent.gz | 124.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5tcm.json.gz 5tcm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tc/5tcm https://data.pdbj.org/pub/pdb/validation_reports/tc/5tcm ftp://data.pdbj.org/pub/pdb/validation_reports/tc/5tcm ftp://data.pdbj.org/pub/pdb/validation_reports/tc/5tcm | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5tckS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 14655.258 Da / Num. of mol.: 3 / Fragment: Bromodomain, UNP residues 8-119 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Leishmania donovani (strain BPK282A1) (eukaryote) Leishmania donovani (strain BPK282A1) (eukaryote)Strain: BPK282A1 / Gene: LDBPK_091320 / Plasmid: PET15-MHL / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): BL21-CodonPlus(DE3)-RIL / References: UniProt: E9BA17 ESCHERICHIA COLI (E. coli) / Strain (production host): BL21-CodonPlus(DE3)-RIL / References: UniProt: E9BA17#2: Chemical | #3: Chemical |  Ethylene glycol Ethylene glycol#4: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.8 Å3/Da / Density % sol: 61.25 % / Mosaicity: 0.836 ° |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, sitting drop / pH: 7 Details: The protein was crystallized at 293 K with 2M NaFormate, 0.1M BTP 7.0 and 1.0 M Cesium chloride with BI2536 using the sitting drop method. |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 19-ID / Wavelength: 0.9788 Å / Beamline: 19-ID / Wavelength: 0.9788 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: DECTRIS PILATUS3 S 6M / Detector: PIXEL / Date: May 2, 2016 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 0.9788 Å / Relative weight: 1 : 0.9788 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.2→50 Å / Num. obs: 24639 / % possible obs: 99 % / Redundancy: 4.5 % / Biso Wilson estimate: 33.15 Å2 / Rmerge(I) obs: 0.132 / Rpim(I) all: 0.065 / Rrim(I) all: 0.148 / Χ2: 1.07 / Net I/av σ(I): 12.58 / Net I/σ(I): 4.8 / Num. measured all: 110927 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
-Phasing
Phasing | Method:  molecular replacement molecular replacement |
|---|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5TCK Resolution: 2.2→44.05 Å / Cor.coef. Fo:Fc: 0.924 / Cor.coef. Fo:Fc free: 0.901 / SU R Cruickshank DPI: 0.554 / Cross valid method: THROUGHOUT / σ(F): 0 / SU R Blow DPI: 0.203 / SU Rfree Blow DPI: 0.169 / SU Rfree Cruickshank DPI: 0.169
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 131.42 Å2 / Biso mean: 35.23 Å2 / Biso min: 12.39 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.25 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 2.2→44.05 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.2→2.3 Å / Total num. of bins used: 12
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj