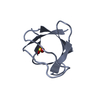
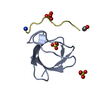
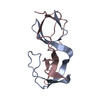
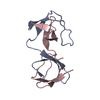
Deposited unit
A: Cytoplasmic protein NCK1
B: Cytoplasmic protein NCK1
C: Cytoplasmic protein NCK1
D: Cytoplasmic protein NCK1
E: Cytoplasmic protein NCK1
F: Cytoplasmic protein NCK1
G: Cytoplasmic protein NCK1
H: Cytoplasmic protein NCK1
I: Cytoplasmic protein NCK1
J: Cytoplasmic protein NCK1
K: Cytoplasmic protein NCK1
L: Cytoplasmic protein NCK1
M: Cytoplasmic protein NCK1
N: Cytoplasmic protein NCK1
O: Cytoplasmic protein NCK1
P: Cytoplasmic protein NCK1
Q: Cytoplasmic protein NCK1
R: Cytoplasmic protein NCK1
S: Cytoplasmic protein NCK1
T: Cytoplasmic protein NCK1
U: Cytoplasmic protein NCK1
V: Cytoplasmic protein NCK1
W: Cytoplasmic protein NCK1
X: Cytoplasmic protein NCK1
Y: Cytoplasmic protein NCK1
Z: Cytoplasmic protein NCK1
1: Cytoplasmic protein NCK1
2: Cytoplasmic protein NCK1 Summary Component details
Theoretical mass Number of molelcules Total (without water) 288,383 28 Polymers 288,383 28 Non-polymers 0 0 Water 3,837 213
1
A: Cytoplasmic protein NCK1
C: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 4070 Å2 ΔGint -21 kcal/mol Surface area 7030 Å2 Method
2
B: Cytoplasmic protein NCK1
D: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 4100 Å2 ΔGint -25 kcal/mol Surface area 6910 Å2 Method
3
E: Cytoplasmic protein NCK1
F: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 4200 Å2 ΔGint -25 kcal/mol Surface area 7140 Å2 Method
4
G: Cytoplasmic protein NCK1
H: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 4170 Å2 ΔGint -23 kcal/mol Surface area 7190 Å2 Method
5
I: Cytoplasmic protein NCK1
J: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 4030 Å2 ΔGint -27 kcal/mol Surface area 7100 Å2 Method
6
K: Cytoplasmic protein NCK1
L: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 4110 Å2 ΔGint -22 kcal/mol Surface area 7310 Å2 Method
7
M: Cytoplasmic protein NCK1
N: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 4040 Å2 ΔGint -23 kcal/mol Surface area 6760 Å2 Method
8
O: Cytoplasmic protein NCK1
P: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 4090 Å2 ΔGint -23 kcal/mol Surface area 7070 Å2 Method
9
Q: Cytoplasmic protein NCK1
R: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 4190 Å2 ΔGint -21 kcal/mol Surface area 6750 Å2 Method
10
S: Cytoplasmic protein NCK1
T: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 4200 Å2 ΔGint -25 kcal/mol Surface area 6930 Å2 Method
11
U: Cytoplasmic protein NCK1
V: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 4040 Å2 ΔGint -20 kcal/mol Surface area 6740 Å2 Method
12
W: Cytoplasmic protein NCK1
X: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 36 2
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 3980 Å2 ΔGint -24 kcal/mol Surface area 6900 Å2 Method
13
Y: Cytoplasmic protein NCK1
Z: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 0
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 3690 Å2 ΔGint -24 kcal/mol Surface area 6880 Å2 Method
14
1: Cytoplasmic protein NCK1
2: Cytoplasmic protein NCK1 Summary Component details Symmetry operations Calculated values
Theoretical mass Number of molelcules Total (without water) 20,599 2 Polymers 20,599 2 Non-polymers 0 0 Water 0
Type Name Symmetry operation Number identity operation 1_555 x,y,z 1
Buried area 3610 Å2 ΔGint -22 kcal/mol Surface area 6330 Å2 Method
Unit cell Length a, b, c (Å) 49.083, 92.053, 114.748 Angle α, β, γ (deg.) 103.020, 91.630, 88.570 Int Tables number 1 Space group name H-M P1
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Cytoplasm
Cytoplasm  Keywords
Keywords SIGNALING PROTEIN /
SIGNALING PROTEIN /  SH3 DOMAIN /
SH3 DOMAIN /  ADAPTOR /
ADAPTOR /  PEPTIDE BINDING / DOMAIN SWAP
PEPTIDE BINDING / DOMAIN SWAP Function and homology information
Function and homology information lamellipodium assembly / RHOV GTPase cycle / positive regulation of actin filament polymerization / negative regulation of PERK-mediated unfolded protein response /
lamellipodium assembly / RHOV GTPase cycle / positive regulation of actin filament polymerization / negative regulation of PERK-mediated unfolded protein response /  protein kinase inhibitor activity / antiviral innate immune response / RHOU GTPase cycle / Generation of second messenger molecules / RHO GTPases Activate WASPs and WAVEs / ephrin receptor signaling pathway / signaling adaptor activity / negative regulation of peptidyl-serine phosphorylation / positive regulation of T cell proliferation /
protein kinase inhibitor activity / antiviral innate immune response / RHOU GTPase cycle / Generation of second messenger molecules / RHO GTPases Activate WASPs and WAVEs / ephrin receptor signaling pathway / signaling adaptor activity / negative regulation of peptidyl-serine phosphorylation / positive regulation of T cell proliferation /  regulation of cell migration /
regulation of cell migration /  T cell activation / negative regulation of insulin receptor signaling pathway / response to endoplasmic reticulum stress /
T cell activation / negative regulation of insulin receptor signaling pathway / response to endoplasmic reticulum stress /  ephrin receptor binding / Downstream signal transduction / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / molecular condensate scaffold activity / actin filament organization / FCGR3A-mediated phagocytosis / PKR-mediated signaling /
ephrin receptor binding / Downstream signal transduction / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / molecular condensate scaffold activity / actin filament organization / FCGR3A-mediated phagocytosis / PKR-mediated signaling /  receptor tyrosine kinase binding / Regulation of actin dynamics for phagocytic cup formation / VEGFA-VEGFR2 Pathway / positive regulation of neuron projection development / cell-cell junction / signaling receptor complex adaptor activity /
receptor tyrosine kinase binding / Regulation of actin dynamics for phagocytic cup formation / VEGFA-VEGFR2 Pathway / positive regulation of neuron projection development / cell-cell junction / signaling receptor complex adaptor activity /  cell migration / protein-macromolecule adaptor activity / Potential therapeutics for SARS /
cell migration / protein-macromolecule adaptor activity / Potential therapeutics for SARS /  ribosome /
ribosome /  cadherin binding / protein domain specific binding /
cadherin binding / protein domain specific binding /  signaling receptor binding / negative regulation of transcription by RNA polymerase II /
signaling receptor binding / negative regulation of transcription by RNA polymerase II /  endoplasmic reticulum /
endoplasmic reticulum /  signal transduction / positive regulation of transcription by RNA polymerase II /
signal transduction / positive regulation of transcription by RNA polymerase II /  nucleus /
nucleus /  plasma membrane /
plasma membrane /  cytosol /
cytosol /  cytoplasm
cytoplasm
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.816 Å
MOLECULAR REPLACEMENT / Resolution: 1.816 Å  Authors
Authors Citation
Citation Journal: J.Biol.Chem. / Year: 2020
Journal: J.Biol.Chem. / Year: 2020 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5qu6.cif.gz
5qu6.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5qu6.ent.gz
pdb5qu6.ent.gz PDB format
PDB format 5qu6.json.gz
5qu6.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/qu/5qu6
https://data.pdbj.org/pub/pdb/validation_reports/qu/5qu6 ftp://data.pdbj.org/pub/pdb/validation_reports/qu/5qu6
ftp://data.pdbj.org/pub/pdb/validation_reports/qu/5qu6 Links
Links Assembly
Assembly














 Components
Components Cytoplasm / NCK adaptor protein 1 / Nck-1 / SH2/SH3 adaptor protein NCK-alpha
Cytoplasm / NCK adaptor protein 1 / Nck-1 / SH2/SH3 adaptor protein NCK-alpha
 Homo sapiens (human) / Gene: NCK1, NCK / Plasmid: PET28a(+) / Production host:
Homo sapiens (human) / Gene: NCK1, NCK / Plasmid: PET28a(+) / Production host: 
 Escherichia coli (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: P16333
Escherichia coli (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: P16333 Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X10SA / Wavelength: 1 Å
/ Beamline: X10SA / Wavelength: 1 Å : 1 Å / Relative weight: 1
: 1 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller
















 PDBj
PDBj

















