| 登録情報 | データベース: PDB / ID: 5gj4
|
|---|
| タイトル | Structure of NS2B-NS3 Protease from Zika Virus caught after self-cleavage |
|---|
 要素 要素 | - Serine protease NS3
- Serine protease subunit NS2B
|
|---|
 キーワード キーワード | HYDROLASE / Zika virus protease / antiviral drug discovery |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / ribonucleoside triphosphate phosphatase activity / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / symbiont-mediated suppression of host toll-like receptor signaling pathway ...symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / ribonucleoside triphosphate phosphatase activity / viral capsid / double-stranded RNA binding / nucleoside-triphosphate phosphatase / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / symbiont-mediated suppression of host toll-like receptor signaling pathway / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / clathrin-dependent endocytosis of virus by host cell / mRNA (nucleoside-2'-O-)-methyltransferase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / protein dimerization activity / symbiont-mediated suppression of host innate immune response / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / RNA helicase / symbiont entry into host cell / induction by virus of host autophagy / serine-type endopeptidase activity / RNA-directed RNA polymerase / viral RNA genome replication / virus-mediated perturbation of host defense response / RNA-dependent RNA polymerase activity / fusion of virus membrane with host endosome membrane / lipid binding / viral envelope / host cell nucleus / virion attachment to host cell / GTP binding / structural molecule activity / virion membrane / ATP hydrolysis activity / proteolysis / extracellular region / ATP binding / membrane / metal ion binding類似検索 - 分子機能 Thrombin, subunit H - #120 / : / Flavivirus capsid protein C superfamily / RNA-directed RNA polymerase, thumb domain, Flavivirus / Flavivirus RNA-directed RNA polymerase, thumb domain / Flavivirus non-structural protein NS2B / Flavivirus envelope glycoprotein E, stem/anchor domain / : / Flavivirus NS3 helicase, C-terminal helical domain / Genome polyprotein, Flavivirus ...Thrombin, subunit H - #120 / : / Flavivirus capsid protein C superfamily / RNA-directed RNA polymerase, thumb domain, Flavivirus / Flavivirus RNA-directed RNA polymerase, thumb domain / Flavivirus non-structural protein NS2B / Flavivirus envelope glycoprotein E, stem/anchor domain / : / Flavivirus NS3 helicase, C-terminal helical domain / Genome polyprotein, Flavivirus / Flavivirus non-structural protein NS4A / Flavivirus non-structural protein NS2B / Flavivirus capsid protein C / Flavivirus non-structural protein NS4B / mRNA cap 0/1 methyltransferase / Flavivirus capsid protein C / Flavivirus non-structural protein NS4B / Flavivirus non-structural protein NS4A / Flavivirus NS2B domain profile. / mRNA cap 0 and cap 1 methyltransferase (EC 2.1.1.56 and EC 2.1.1.57) domain profile. / Flavivirus non-structural protein NS2A / Flavivirus non-structural protein NS2A / Flavivirus NS3, petidase S7 / Peptidase S7, Flavivirus NS3 serine protease / Flavivirus NS3 protease (NS3pro) domain profile. / RNA-directed RNA polymerase, flavivirus / Flavivirus RNA-directed RNA polymerase, fingers and palm domains / Flavivirus non-structural Protein NS1 / Flavivirus non-structural protein NS1 / Envelope glycoprotein M, flavivirus / Flavivirus envelope glycoprotein M / Envelope glycoprotein M superfamily, flavivirus / Flavivirus polyprotein propeptide / Flavivirus polyprotein propeptide superfamily / Flavivirus polyprotein propeptide / Flaviviral glycoprotein E, central domain, subdomain 1 / Flaviviral glycoprotein E, central domain, subdomain 2 / Flavivirus envelope glycoprotein E, Stem/Anchor domain / Flavivirus glycoprotein E, immunoglobulin-like domain / Flavivirus envelope glycoprotein E, Stem/Anchor domain superfamily / Flavivirus glycoprotein, immunoglobulin-like domain / Flavivirus glycoprotein central and dimerisation domain / Flavivirus glycoprotein, central and dimerisation domains / Ribosomal RNA methyltransferase, FtsJ domain / FtsJ-like methyltransferase / Flavivirus/Alphavirus glycoprotein, immunoglobulin-like domain superfamily / Flavivirus glycoprotein, central and dimerisation domain superfamily / Flaviviral glycoprotein E, dimerisation domain / DEAD box, Flavivirus / Flavivirus DEAD domain / helicase superfamily c-terminal domain / Trypsin-like serine proteases / Immunoglobulin E-set / Superfamilies 1 and 2 helicase C-terminal domain profile. / Superfamilies 1 and 2 helicase ATP-binding type-1 domain profile. / DEAD-like helicases superfamily / Helicase, C-terminal / Helicase superfamily 1/2, ATP-binding domain / Thrombin, subunit H / RNA-directed RNA polymerase, catalytic domain / RdRp of positive ssRNA viruses catalytic domain profile. / S-adenosyl-L-methionine-dependent methyltransferase superfamily / Peptidase S1, PA clan / DNA/RNA polymerase superfamily / Beta Barrel / P-loop containing nucleoside triphosphate hydrolase / Mainly Beta類似検索 - ドメイン・相同性 |
|---|
| 生物種 |   Zika virus (ジカ熱ウイルス) Zika virus (ジカ熱ウイルス) |
|---|
| 手法 |  X線回折 / X線回折 /  シンクロトロン / シンクロトロン /  分子置換 / 解像度: 1.839 Å 分子置換 / 解像度: 1.839 Å |
|---|
 データ登録者 データ登録者 | Phoo, W.W. / Li, Y. |
|---|
| 資金援助 |  シンガポール, 1件 シンガポール, 1件 | 組織 | 認可番号 | 国 |
|---|
| NMRC | CBRG14May051 |  シンガポール シンガポール |
|
|---|
 引用 引用 |  ジャーナル: Nat Commun / 年: 2016 ジャーナル: Nat Commun / 年: 2016
タイトル: Structure of the NS2B-NS3 protease from Zika virus after self-cleavage.
著者: Phoo, W.W. / Li, Y. / Zhang, Z. / Lee, M.Y. / Loh, Y.R. / Tan, Y.B. / Ng, E.Y. / Lescar, J. / Kang, C. / Luo, D. |
|---|
| 履歴 | | 登録 | 2016年6月27日 | 登録サイト: PDBJ / 処理サイト: PDBJ |
|---|
| 改定 1.0 | 2016年10月26日 | Provider: repository / タイプ: Initial release |
|---|
| 改定 1.1 | 2016年12月7日 | Group: Database references |
|---|
| 改定 1.2 | 2023年11月8日 | Group: Data collection / Database references / Refinement description
カテゴリ: chem_comp_atom / chem_comp_bond ...chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model
Item: _database_2.pdbx_DOI / _database_2.pdbx_database_accession |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報
 Zika virus (ジカ熱ウイルス)
Zika virus (ジカ熱ウイルス) X線回折 /
X線回折 /  シンクロトロン /
シンクロトロン /  分子置換 / 解像度: 1.839 Å
分子置換 / 解像度: 1.839 Å  データ登録者
データ登録者 シンガポール, 1件
シンガポール, 1件  引用
引用 ジャーナル: Nat Commun / 年: 2016
ジャーナル: Nat Commun / 年: 2016 構造の表示
構造の表示 Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 5gj4.cif.gz
5gj4.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb5gj4.ent.gz
pdb5gj4.ent.gz PDB形式
PDB形式 5gj4.json.gz
5gj4.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 5gj4_validation.pdf.gz
5gj4_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 5gj4_full_validation.pdf.gz
5gj4_full_validation.pdf.gz 5gj4_validation.xml.gz
5gj4_validation.xml.gz 5gj4_validation.cif.gz
5gj4_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/gj/5gj4
https://data.pdbj.org/pub/pdb/validation_reports/gj/5gj4 ftp://data.pdbj.org/pub/pdb/validation_reports/gj/5gj4
ftp://data.pdbj.org/pub/pdb/validation_reports/gj/5gj4
 リンク
リンク 集合体
集合体

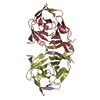
 要素
要素
 Zika virus (strain Mr 766) (ジカ熱ウイルス)
Zika virus (strain Mr 766) (ジカ熱ウイルス)

 Zika virus (strain Mr 766) (ジカ熱ウイルス)
Zika virus (strain Mr 766) (ジカ熱ウイルス)
 X線回折 / 使用した結晶の数: 1
X線回折 / 使用した結晶の数: 1  試料調製
試料調製 シンクロトロン / サイト:
シンクロトロン / サイト:  SLS
SLS  / ビームライン: X06SA / 波長: 1.0004 Å
/ ビームライン: X06SA / 波長: 1.0004 Å 解析
解析 分子置換
分子置換 ムービー
ムービー コントローラー
コントローラー













 PDBj
PDBj




