[English] 日本語
 Yorodumi
Yorodumi- PDB-5c9m: The structure of oxidized rat cytochrome c (T28A) at 1.362 angstr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5c9m | ||||||
|---|---|---|---|---|---|---|---|
| Title | The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution. | ||||||
 Components Components | Cytochrome c, somatic | ||||||
 Keywords Keywords |  ELECTRON TRANSPORT / cytochrome c oxidized rat mutant ELECTRON TRANSPORT / cytochrome c oxidized rat mutant | ||||||
| Function / homology |  Function and homology information Function and homology informationRelease of apoptotic factors from the mitochondria / Formation of apoptosome / Activation of caspases through apoptosome-mediated cleavage / Transcriptional activation of mitochondrial biogenesis /  Pyroptosis / Respiratory electron transport / Detoxification of Reactive Oxygen Species / TP53 Regulates Metabolic Genes / response to carbon monoxide / Cytoprotection by HMOX1 ...Release of apoptotic factors from the mitochondria / Formation of apoptosome / Activation of caspases through apoptosome-mediated cleavage / Transcriptional activation of mitochondrial biogenesis / Pyroptosis / Respiratory electron transport / Detoxification of Reactive Oxygen Species / TP53 Regulates Metabolic Genes / response to carbon monoxide / Cytoprotection by HMOX1 ...Release of apoptotic factors from the mitochondria / Formation of apoptosome / Activation of caspases through apoptosome-mediated cleavage / Transcriptional activation of mitochondrial biogenesis /  Pyroptosis / Respiratory electron transport / Detoxification of Reactive Oxygen Species / TP53 Regulates Metabolic Genes / response to carbon monoxide / Cytoprotection by HMOX1 / Pyroptosis / Respiratory electron transport / Detoxification of Reactive Oxygen Species / TP53 Regulates Metabolic Genes / response to carbon monoxide / Cytoprotection by HMOX1 /  apoptosome / Regulation of the apoptosome activity / negative regulation of hydrogen peroxide biosynthetic process / response to gravity / positive regulation of cellular respiration / glial cell apoptotic process / response to copper ion / mitochondrial electron transport, cytochrome c to oxygen / mitochondrial electron transport, ubiquinol to cytochrome c / hydrogen peroxide metabolic process / apoptosome / Regulation of the apoptosome activity / negative regulation of hydrogen peroxide biosynthetic process / response to gravity / positive regulation of cellular respiration / glial cell apoptotic process / response to copper ion / mitochondrial electron transport, cytochrome c to oxygen / mitochondrial electron transport, ubiquinol to cytochrome c / hydrogen peroxide metabolic process /  respirasome / response to ischemia / respirasome / response to ischemia /  mitochondrial intermembrane space / response to oxidative stress / mitochondrial intermembrane space / response to oxidative stress /  electron transfer activity / apoptotic process / electron transfer activity / apoptotic process /  heme binding / heme binding /  enzyme binding / enzyme binding /  mitochondrion / mitochondrion /  metal ion binding / metal ion binding /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.362 Å MOLECULAR REPLACEMENT / Resolution: 1.362 Å | ||||||
 Authors Authors | Edwards, B.F.P. / Mahapatra, G. / Vaishnav, A.A. / Brunzelle, J.S. / Huttemann, M. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: The structure of oxidized rat cytochrome c (T28A) at 1.362 angstroms resolution. Authors: Edwards, B.F.P. / Mahapatra, G. / Vaishnav, A.A. / Brunzelle, J.S. / Huttemann, M. #1: Journal: Biochemistry / Year: 2010 Title: Phosphomimetic substitution of cytochrome C tyrosine 48 decreases respiration and binding to cardiolipin and abolishes ability to trigger downstream caspase activation. Authors: Pecina, P. / Borisenko, G.G. / Belikova, N.A. / Tyurina, Y.Y. / Pecinova, A. / Lee, I. / Samhan-Arias, A.K. / Przyklenk, K. / Kagan, V.E. / Huttemann, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
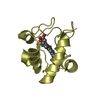
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5c9m.cif.gz 5c9m.cif.gz | 205.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5c9m.ent.gz pdb5c9m.ent.gz | 165.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5c9m.json.gz 5c9m.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c9/5c9m https://data.pdbj.org/pub/pdb/validation_reports/c9/5c9m ftp://data.pdbj.org/pub/pdb/validation_reports/c9/5c9m ftp://data.pdbj.org/pub/pdb/validation_reports/c9/5c9m | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5c0zS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | Monomer confirmed by gel filtration |
- Components
Components
| #1: Protein |  Mass: 11598.462 Da / Num. of mol.: 4 / Mutation: T29A Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Rattus norvegicus (Norway rat) / Tissue: somatic / Gene: Cycs / Plasmid: pLW01 / Cell line (production host): C41 (DE3) / Production host: Rattus norvegicus (Norway rat) / Tissue: somatic / Gene: Cycs / Plasmid: pLW01 / Cell line (production host): C41 (DE3) / Production host:   Escherichia coli (E. coli) / References: UniProt: P62898 Escherichia coli (E. coli) / References: UniProt: P62898#2: Chemical | ChemComp-HEC /  Heme C Heme C#3: Chemical | ChemComp-FC6 / #4: Water | ChemComp-HOH / |  Water WaterSequence details | Mouse Cytochrome c has the same sequence (UNP P62897/CYC_MOUSE) as this entry (UNP P62898/CYC_RAT) | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.27 Å3/Da / Density % sol: 45.7 % / Description: 0.3 x0.1 mm prisms |
|---|---|
Crystal grow | Temperature: 295 K / Method: vapor diffusion, sitting drop / pH: 6.5 Details: 20 mg/ml protein, PEG 4000, isopropanol, sodium acetate, potassium ferricyanate PH range: 6.5-7.0 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 21-ID-F / Wavelength: 0.97872 Å / Beamline: 21-ID-F / Wavelength: 0.97872 Å |
| Detector | Type: MARMOSAIC 225 mm CCD / Detector: CCD / Date: Nov 18, 2014 |
| Radiation | Monochromator: diamond laue / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.97872 Å / Relative weight: 1 : 0.97872 Å / Relative weight: 1 |
| Reflection | Resolution: 1.362→57.962 Å / Num. all: 80448 / Num. obs: 76544 / % possible obs: 92.37 % / Redundancy: 3.9 % / Biso Wilson estimate: 13 Å2 / Rmerge(I) obs: 0.052 / Net I/σ(I): 14.1 |
| Reflection shell | Resolution: 1.362→1.398 Å / Redundancy: 3.8 % / Rmerge(I) obs: 0.58 / Mean I/σ(I) obs: 2.2 / % possible all: 66.24 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 5C0Z Resolution: 1.362→57.96 Å / Cor.coef. Fo:Fc: 0.979 / Cor.coef. Fo:Fc free: 0.969 / SU B: 2.244 / SU ML: 0.039 / Cross valid method: THROUGHOUT / ESU R: 0.057 / ESU R Free: 0.055 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.7 Å / Shrinkage radii: 0.7 Å / VDW probe radii: 1 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 21.413 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.362→57.96 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj












