+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5bvi | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | X-ray Structure of Interferon Regulatory Factor 4 IAD Domain | ||||||||||||
 Components Components | Interferon regulatory factor 4 Interferon regulatory factors Interferon regulatory factors | ||||||||||||
 Keywords Keywords |  TRANSCRIPTION / TRANSCRIPTION /  Interferon Regulatory Factors / Transcription activation/Protein-DNA interaction Interferon Regulatory Factors / Transcription activation/Protein-DNA interaction | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationnucleosome => GO:0000786 / : / : / negative regulation of toll-like receptor signaling pathway / T-helper 17 cell lineage commitment / myeloid dendritic cell differentiation / positive regulation of interleukin-13 production /  immune system process / defense response to protozoan / positive regulation of interleukin-10 production ...nucleosome => GO:0000786 / : / : / negative regulation of toll-like receptor signaling pathway / T-helper 17 cell lineage commitment / myeloid dendritic cell differentiation / positive regulation of interleukin-13 production / immune system process / defense response to protozoan / positive regulation of interleukin-10 production ...nucleosome => GO:0000786 / : / : / negative regulation of toll-like receptor signaling pathway / T-helper 17 cell lineage commitment / myeloid dendritic cell differentiation / positive regulation of interleukin-13 production /  immune system process / defense response to protozoan / positive regulation of interleukin-10 production / positive regulation of interleukin-4 production / positive regulation of DNA binding / positive regulation of interleukin-2 production / sequence-specific double-stranded DNA binding / positive regulation of cold-induced thermogenesis / DNA-binding transcription activator activity, RNA polymerase II-specific / sequence-specific DNA binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / positive regulation of DNA-templated transcription / positive regulation of transcription by RNA polymerase II / immune system process / defense response to protozoan / positive regulation of interleukin-10 production / positive regulation of interleukin-4 production / positive regulation of DNA binding / positive regulation of interleukin-2 production / sequence-specific double-stranded DNA binding / positive regulation of cold-induced thermogenesis / DNA-binding transcription activator activity, RNA polymerase II-specific / sequence-specific DNA binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / positive regulation of DNA-templated transcription / positive regulation of transcription by RNA polymerase II /  nucleoplasm / nucleoplasm /  nucleus / nucleus /  cytosol cytosolSimilarity search - Function | ||||||||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | ||||||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.6 Å MOLECULAR REPLACEMENT / Resolution: 2.6 Å | ||||||||||||
 Authors Authors | Escalate, C.R. / Remesh, S.G. | ||||||||||||
| Funding support |  United States, 3items United States, 3items
| ||||||||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2015 Journal: J.Biol.Chem. / Year: 2015Title: Structural Studies of IRF4 Reveal a Flexible Autoinhibitory Region and a Compact Linker Domain. Authors: Remesh, S.G. / Santosh, V. / Escalante, C.R. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5bvi.cif.gz 5bvi.cif.gz | 87.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5bvi.ent.gz pdb5bvi.ent.gz | 65 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5bvi.json.gz 5bvi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bv/5bvi https://data.pdbj.org/pub/pdb/validation_reports/bv/5bvi ftp://data.pdbj.org/pub/pdb/validation_reports/bv/5bvi ftp://data.pdbj.org/pub/pdb/validation_reports/bv/5bvi | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3dshS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 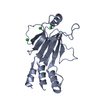
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | Monomer confirmed by analytical ultracentrifugation |
- Components
Components
| #1: Protein |  Interferon regulatory factors / Interferon regulatory factor 4 / isoform CRA_b Interferon regulatory factors / Interferon regulatory factor 4 / isoform CRA_bMass: 21463.451 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Gene: Irf4, mCG_4922 / Plasmid: pet15TEV_NESG Mus musculus (house mouse) / Gene: Irf4, mCG_4922 / Plasmid: pet15TEV_NESGDetails (production host): EvNO00338203 (pet15b with TEV site) Production host:   Escherichia coli (E. coli) / Strain (production host): BL21(DE3)pLysS star / References: UniProt: Q5SUZ4, UniProt: Q64287*PLUS Escherichia coli (E. coli) / Strain (production host): BL21(DE3)pLysS star / References: UniProt: Q5SUZ4, UniProt: Q64287*PLUS#2: Chemical | ChemComp-CL /  Chloride Chloride#3: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.46 Å3/Da / Density % sol: 64.42 % |
|---|---|
Crystal grow | Temperature: 277.15 K / Method: vapor diffusion, hanging drop / Details: 1.5-1.7M KCl, 0.1M Imidazole pH 8.0 |
-Data collection
| Diffraction | Mean temperature: 193.15 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  NSLS NSLS  / Beamline: X25 / Wavelength: 1.1 Å / Beamline: X25 / Wavelength: 1.1 Å |
| Detector | Type: PSI PILATUS 6M / Detector: PIXEL / Date: Oct 18, 2013 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.1 Å / Relative weight: 1 : 1.1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.36→50 Å / Num. obs: 23107 / % possible obs: 93.4 % / Redundancy: 4 % / Rmerge(I) obs: 0.153 / Net I/σ(I): 8.7 |
| Reflection shell | Resolution: 2.36→2.4 Å / Redundancy: 3.1 % / Rmerge(I) obs: 0.66 / Mean I/σ(I) obs: 2.1 / % possible all: 89.1 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 3DSH Resolution: 2.6→32.337 Å / SU ML: 0.33 / Cross valid method: FREE R-VALUE / σ(F): 1.37 / Phase error: 29.65 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.6→32.337 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj




