+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4uzs | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of Bifidobacterium bifidum beta-galactosidase | ||||||
 Components Components | BETA-GALACTOSIDASE | ||||||
 Keywords Keywords |  HYDROLASE / HYDROLASE /  LACTASE / FAMILY 42 LACTASE / FAMILY 42 | ||||||
| Function / homology |  Function and homology information Function and homology informationgalactose metabolic process /  beta-galactosidase complex / beta-galactosidase complex /  beta-galactosidase / beta-galactosidase /  beta-galactosidase activity beta-galactosidase activitySimilarity search - Function | ||||||
| Biological species |   BIFIDOBACTERIUM BIFIDUM S17 (bacteria) BIFIDOBACTERIUM BIFIDUM S17 (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SAD / Resolution: 1.74 Å SAD / Resolution: 1.74 Å | ||||||
 Authors Authors | Godoy, A.S. / Murakami, M.T. / Camilo, C.M. / Bernardes, A. / Polikarpov, I. | ||||||
 Citation Citation |  Journal: FEBS J. / Year: 2016 Journal: FEBS J. / Year: 2016Title: Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpah-Galactose. Authors: Godoy, A.S. / Camilo, C.M. / Kadowaki, M.A. / Muniz, H.D.S. / Santo, M.E. / Murakami, M.T. / Nascimento, A.S. / Polikarpov, I. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4uzs.cif.gz 4uzs.cif.gz | 466 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4uzs.ent.gz pdb4uzs.ent.gz | 381.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4uzs.json.gz 4uzs.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/uz/4uzs https://data.pdbj.org/pub/pdb/validation_reports/uz/4uzs ftp://data.pdbj.org/pub/pdb/validation_reports/uz/4uzs ftp://data.pdbj.org/pub/pdb/validation_reports/uz/4uzs | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 3 molecules ABC
| #1: Protein |  / BETA-GAL / BETA-GALMass: 77278.234 Da / Num. of mol.: 3 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   BIFIDOBACTERIUM BIFIDUM S17 (bacteria) BIFIDOBACTERIUM BIFIDUM S17 (bacteria)Description: COURTESY OF PROF. CHRISTIAN RIEDEL, ULM UNIVERSITY, GERMANY Plasmid: PPROEX HTA / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): BL21 / References: UniProt: E3EPA1, ESCHERICHIA COLI (E. coli) / Strain (production host): BL21 / References: UniProt: E3EPA1,  beta-galactosidase beta-galactosidase |
|---|
-Non-polymers , 5 types, 2713 molecules 


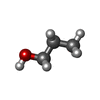





| #2: Chemical |  Diethylene glycol Diethylene glycol#3: Chemical | ChemComp-GOL /  Glycerol Glycerol#4: Chemical | ChemComp-P6G / |  Polyethylene glycol Polyethylene glycol#5: Chemical | ChemComp-POL /  Propan-1-ol Propan-1-ol#6: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.35 Å3/Da / Density % sol: 47.7 % / Description: CRYSTALS DERIVATED WITH HG |
|---|---|
Crystal grow | Temperature: 292 K / pH: 6 Details: 20% (W/V) POLYETHYLENE GLYCOL 3, 350, 0.2 M DIBASIC AMMONIUM TARTRATE AND 4% (W/V) 1-PROPANOL AT 292 K, pH 6 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.54 ROTATING ANODE / Type: RIGAKU MICROMAX-007 / Wavelength: 1.54 |
| Detector | Type: MARRESEARCH / Detector: IMAGE PLATE / Date: Jun 6, 2010 / Details: MIRROR |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.54 Å / Relative weight: 1 : 1.54 Å / Relative weight: 1 |
| Reflection | Resolution: 1.7→35.7 Å / Num. obs: 200160 / % possible obs: 98.5 % / Observed criterion σ(I): 1.8 / Redundancy: 3.1 % / Biso Wilson estimate: 16.8 Å2 / Rmerge(I) obs: 0.09 / Net I/σ(I): 11.8 |
| Reflection shell | Resolution: 1.7→1.8 Å / Redundancy: 2.7 % / Rmerge(I) obs: 0.65 / Mean I/σ(I) obs: 1.8 / % possible all: 89.9 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  SAD SADStarting model: NONE Resolution: 1.74→35.772 Å / SU ML: 0.2 / σ(F): 1.34 / Phase error: 20.59 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.74→35.772 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj







