[English] 日本語
 Yorodumi
Yorodumi- PDB-4l26: Crystal structure of DNA duplex containing consecutive T-T mispai... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4l26 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Crystal structure of DNA duplex containing consecutive T-T mispairs (Br-derivative) | ||||||||||||||||||
 Components Components | DNA (5'-D(* Keywords Keywords DNA / T-T mispair / A-T-T triplet / non-helical DNA duplex DNA / T-T mispair / A-T-T triplet / non-helical DNA duplexFunction / homology |  DNA / DNA (> 10) DNA / DNA (> 10) Function and homology information Function and homology informationMethod |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MAD / Resolution: 1.4 Å MAD / Resolution: 1.4 Å  Authors AuthorsKondo, J. / Yamada, T. / Hirose, C. / Tanaka, Y. / Ono, A. |  Citation Citation Journal: Angew.Chem.Int.Ed.Engl. / Year: 2014 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2014Title: Crystal Structure of Metallo DNA Duplex Containing Consecutive Watson-Crick-like T-Hg(II) -T Base Pairs Authors: Kondo, J. / Yamada, T. / Hirose, C. / Okamoto, I. / Tanaka, Y. / Ono, A. History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4l26.cif.gz 4l26.cif.gz | 25 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4l26.ent.gz pdb4l26.ent.gz | 16.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4l26.json.gz 4l26.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/l2/4l26 https://data.pdbj.org/pub/pdb/validation_reports/l2/4l26 ftp://data.pdbj.org/pub/pdb/validation_reports/l2/4l26 ftp://data.pdbj.org/pub/pdb/validation_reports/l2/4l26 | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 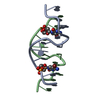
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: DNA chain | Mass: 3733.274 Da / Num. of mol.: 2 / Source method: obtained synthetically / Details: Chemically synthesized #2: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.16 Å3/Da / Density % sol: 43.17 % |
|---|---|
Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 7 Details: Sodium Cacodylate, Spermine tetrahydrochloride, 2-methyl-2,4-pentanediol, ammonium chloride , pH 7.0, VAPOR DIFFUSION, HANGING DROP, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 277 K | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Photon Factory Photon Factory  / Beamline: AR-NW12A / Wavelength: 1.0, 0.91942, 0.92006 / Beamline: AR-NW12A / Wavelength: 1.0, 0.91942, 0.92006 | ||||||||||||
| Detector | Date: Oct 22, 2012 | ||||||||||||
| Radiation | Protocol: MAD / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||
| Radiation wavelength |
| ||||||||||||
| Reflection | Resolution: 1.4→21.94 Å / Num. obs: 12212 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MAD / Resolution: 1.4→21.94 Å / Occupancy max: 1 / Occupancy min: 0.5 / σ(F): 0 MAD / Resolution: 1.4→21.94 Å / Occupancy max: 1 / Occupancy min: 0.5 / σ(F): 0
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Bsol: 52.9486 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 52.41 Å2 / Biso mean: 19.0444 Å2 / Biso min: 9.74 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.4→21.94 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 10
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj







































