+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4hb1 | ||||||
|---|---|---|---|---|---|---|---|
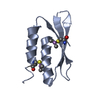
| Title | A DESIGNED FOUR HELIX BUNDLE PROTEIN. | ||||||
 Components Components | DHP1 | ||||||
 Keywords Keywords | DESIGNED HELICAL BUNDLE | ||||||
| Function / homology | Immunoglobulin FC, subunit C / Single alpha-helices involved in coiled-coils or other helix-helix interfaces / Up-down Bundle / Mainly Alpha Function and homology information Function and homology information | ||||||
| Biological species | synthetic construct (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.9 Å MOLECULAR REPLACEMENT / Resolution: 2.9 Å | ||||||
 Authors Authors | Schafmeister, C.E. / Laporte, S.L. / Miercke, L.J.W. / Stroud, R.M. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 1997 Journal: Nat.Struct.Biol. / Year: 1997Title: A designed four helix bundle protein with native-like structure. Authors: Schafmeister, C.E. / LaPorte, S.L. / Miercke, L.J. / Stroud, R.M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4hb1.cif.gz 4hb1.cif.gz | 18.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4hb1.ent.gz pdb4hb1.ent.gz | 11.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4hb1.json.gz 4hb1.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/hb/4hb1 https://data.pdbj.org/pub/pdb/validation_reports/hb/4hb1 ftp://data.pdbj.org/pub/pdb/validation_reports/hb/4hb1 ftp://data.pdbj.org/pub/pdb/validation_reports/hb/4hb1 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | THE MOLECULE CONTAINS A CRYSTALLOGRAPHIC TW0-FOLD AXIS OF SYMMETRY THROUGH THE CENTER OF THE 4-HELIX BUNDLE. THIS UNUSUAL ARRANGEMENT IS DUE TO THE SYNTHETIC NATURE OF THE MOLECULE IN WHICH THE SAME SEQUENCE SEGMENTS REPEAT. THE SYMMETRY OPERATION REQUIRED TO GENERATE THE ENTIRE BUNDLE IS (-Y+1, -X+1, 1/3-Z) AND IS GIVEN IN REMARK 350 AS AN OPERATOR ON THE COORDINATES IN THIS ENTRY. |
- Components
Components
| #1: Protein | Mass: 11815.356 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: DE NOVO DESIGN A 108 AMINO ACID PROTEIN WAS DESIGNED AND CONSTRUCTED FROM A REDUCED ALPHABET OF SEVEN AMINO ACIDS. THE 2.9 ANGSTROM CRYSTAL STRUCTURE CONFIRMS THAT THE PROTEIN IS A FOUR ...Details: DE NOVO DESIGN A 108 AMINO ACID PROTEIN WAS DESIGNED AND CONSTRUCTED FROM A REDUCED ALPHABET OF SEVEN AMINO ACIDS. THE 2.9 ANGSTROM CRYSTAL STRUCTURE CONFIRMS THAT THE PROTEIN IS A FOUR HELIX BUNDLE, AS IT WAS DESIGNED TO BE. THE COMPLETE 108 RESIDUE SEQUENCE IS PRESENTED ON SEQRES RECORDS BELOW. THE HELICES WERE CONNECTED BY LOOPS OF 3, 4, 3 GLYCINES, RESPECTIVELY. THE MOLECULE CONTAINS A CRYSTALLOGRAPHIC TW0-FOLD AXIS OF SYMMETRY THROUGH THE CENTER OF THE 4-HELIX BUNDLE. THIS UNUSUAL ARRANGEMENT IS DUE TO THE SYNTHETIC NATURE OF THE MOLECULE IN WHICH THE SAME SEQUENCE SEGMENTS REPEAT. THE SYMMETRY OPERATION REQUIRED TO GENERATE THE ENTIRE BUNDLE IS (-Y+1, -X+1, 1/3-Z) AND IS GIVEN IN REMARK 350 AS AN OPERATOR ON THE COORDINATES IN THIS ENTRY. Source: (gene. exp.) synthetic construct (others) / Description: DE NOVO DESIGNED FOUR HELIX BUNDLE PROTEIN / Gene: SYNTHETIC GENE DHP1 / Plasmid: PMAL-C2 / Gene (production host): SYNTHETIC GENE DHP1 / Production host:   Escherichia coli (E. coli) / Strain (production host): XL1-BLUE Escherichia coli (E. coli) / Strain (production host): XL1-BLUE | ||||
|---|---|---|---|---|---|
| #2: Chemical | | #3: Water | ChemComp-HOH / |  Water WaterCompound details | A 108 AMINO ACID PROTEIN WAS DESIGNED AND CONSTRUCTED FROM A REDUCED ALPHABET OF SEVEN AMINO ACIDS. ...A 108 AMINO ACID PROTEIN WAS DESIGNED AND CONSTRUCTE | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
Crystal grow | pH: 8.6 Details: 65% AMMONIUM SULPHATE, 3% ISOPROPANOL, 100MM TRIS, PH 8.6 | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | *PLUS Method: vapor diffusion, sitting drop | |||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Type: / Type:  SSRL SSRL  / Wavelength: 1.5418 / Wavelength: 1.5418 |
| Detector | Date: May 1, 1995 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Highest resolution: 2.9 Å / Num. obs: 19263 / % possible obs: 96 % / Observed criterion σ(I): 2 / Redundancy: 8.2 % / Biso Wilson estimate: 60 Å2 / Rsym value: 0.055 |
| Reflection shell | Highest resolution: 2.9 Å |
| Reflection | *PLUS Num. obs: 1212 / Num. measured all: 19263 / Rmerge(I) obs: 0.055 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: MODEL HELIX Resolution: 2.9→7.5 Å / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.9→7.5 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj


