+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4fxx | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of SF1 coiled-coil domain | ||||||
 Components Components | Splicing factor 1 | ||||||
 Keywords Keywords |  RNA BINDING PROTEIN / RNA BINDING PROTEIN /  splicing factor 1 / splicing factor 1 /  coiled-coil / pre-mRNA splicing / U2AF65-UHM binding coiled-coil / pre-mRNA splicing / U2AF65-UHM binding | ||||||
| Function / homology |  Function and homology information Function and homology informationnuclear body organization / U2AF complex / regulation of steroid biosynthetic process / Leydig cell differentiation / male sex determination /  regulation of mRNA splicing, via spliceosome / spliceosomal complex assembly / mRNA 3'-splice site recognition / mRNA Splicing - Major Pathway / negative regulation of smooth muscle cell proliferation ...nuclear body organization / U2AF complex / regulation of steroid biosynthetic process / Leydig cell differentiation / male sex determination / regulation of mRNA splicing, via spliceosome / spliceosomal complex assembly / mRNA 3'-splice site recognition / mRNA Splicing - Major Pathway / negative regulation of smooth muscle cell proliferation ...nuclear body organization / U2AF complex / regulation of steroid biosynthetic process / Leydig cell differentiation / male sex determination /  regulation of mRNA splicing, via spliceosome / spliceosomal complex assembly / mRNA 3'-splice site recognition / mRNA Splicing - Major Pathway / negative regulation of smooth muscle cell proliferation / regulation of mRNA splicing, via spliceosome / spliceosomal complex assembly / mRNA 3'-splice site recognition / mRNA Splicing - Major Pathway / negative regulation of smooth muscle cell proliferation /  spliceosomal complex / spliceosomal complex /  mRNA splicing, via spliceosome / transcription corepressor activity / mRNA splicing, via spliceosome / transcription corepressor activity /  nuclear body / nuclear body /  ribosome / ribosome /  mRNA binding / mRNA binding /  RNA binding / zinc ion binding / RNA binding / zinc ion binding /  nucleoplasm / identical protein binding / nucleoplasm / identical protein binding /  nucleus nucleusSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.4801 Å MOLECULAR REPLACEMENT / Resolution: 2.4801 Å | ||||||
 Authors Authors | Gupta, A. / Bauer, W.J. / Wang, W. / Kielkopf, C.L. | ||||||
 Citation Citation |  Journal: Structure / Year: 2013 Journal: Structure / Year: 2013Title: Structure of Phosphorylated SF1 Bound to U2AF(65) in an Essential Splicing Factor Complex. Authors: Wang, W. / Maucuer, A. / Gupta, A. / Manceau, V. / Thickman, K.R. / Bauer, W.J. / Kennedy, S.D. / Wedekind, J.E. / Green, M.R. / Kielkopf, C.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4fxx.cif.gz 4fxx.cif.gz | 87.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4fxx.ent.gz pdb4fxx.ent.gz | 67.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4fxx.json.gz 4fxx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fx/4fxx https://data.pdbj.org/pub/pdb/validation_reports/fx/4fxx ftp://data.pdbj.org/pub/pdb/validation_reports/fx/4fxx ftp://data.pdbj.org/pub/pdb/validation_reports/fx/4fxx | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 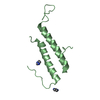
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| 5 | 
| ||||||||
| 6 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  / Mammalian branch point-binding protein / BBP / mBBP / Transcription factor ZFM1 / Zinc finger gene ...Mammalian branch point-binding protein / BBP / mBBP / Transcription factor ZFM1 / Zinc finger gene in MEN1 locus / Zinc finger protein 162 / Mammalian branch point-binding protein / BBP / mBBP / Transcription factor ZFM1 / Zinc finger gene ...Mammalian branch point-binding protein / BBP / mBBP / Transcription factor ZFM1 / Zinc finger gene in MEN1 locus / Zinc finger protein 162Mass: 12762.532 Da / Num. of mol.: 4 / Fragment: UNP Residues 26-132 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: SF1, ZFM1, ZNF162 / Plasmid: pGEX-6p / Production host: Homo sapiens (human) / Gene: SF1, ZFM1, ZNF162 / Plasmid: pGEX-6p / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21 Rosetta / References: UniProt: Q15637 Escherichia coli (E. coli) / Strain (production host): BL21 Rosetta / References: UniProt: Q15637#2: Chemical | ChemComp-IMD /  Imidazole Imidazole#3: Chemical |  Malonic acid Malonic acid#4: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.47 Å3/Da / Density % sol: 50.11 % |
|---|---|
Crystal grow | Temperature: 277 K / Method: vapor diffusion, hanging drop / pH: 5.5 Details: 1.34 M sodium malonate pH 6.0, 0.1 M imidazole maleate pH 5.5, VAPOR DIFFUSION, HANGING DROP, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 100 K | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-2 / Wavelength: 0.9794 Å / Beamline: BL9-2 / Wavelength: 0.9794 Å | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: MARMOSAIC 325 mm CCD / Detector: CCD / Date: Dec 15, 2011 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 0.9794 Å / Relative weight: 1 : 0.9794 Å / Relative weight: 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.48→48 Å / Num. all: 18175 / Num. obs: 16211 / % possible obs: 89.19 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 5.08 % / Rmerge(I) obs: 0.108 / Net I/σ(I): 10.5645 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell |
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT / Resolution: 2.4801→34.966 Å / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.7967 / SU ML: 0.87 / σ(F): 0 / Phase error: 27.29 / Stereochemistry target values: ML MOLECULAR REPLACEMENT / Resolution: 2.4801→34.966 Å / Occupancy max: 1 / Occupancy min: 0.5 / FOM work R set: 0.7967 / SU ML: 0.87 / σ(F): 0 / Phase error: 27.29 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.72 Å / VDW probe radii: 1 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 53.151 Å2 / ksol: 0.379 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 117.25 Å2 / Biso mean: 46.2666 Å2 / Biso min: 11.78 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.4801→34.966 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION / Total num. of bins used: 9
|
 Movie
Movie Controller
Controller














 PDBj
PDBj












