+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4fmx | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal Structure of Substrate-Bound P450cin | ||||||
 Components Components | P450cin 1,8-Cineole 2-endo-monooxygenase 1,8-Cineole 2-endo-monooxygenase | ||||||
 Keywords Keywords |  OXIDOREDUCTASE / OXIDOREDUCTASE /  P450 / P450 /  HEME / HEME /  MONOOXYGENASE / CINDOXIN MONOOXYGENASE / CINDOXIN | ||||||
| Function / homology |  Function and homology information Function and homology information 1,8-cineole 2-endo-monooxygenase / carbazole catabolic process / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen / iron ion binding / 1,8-cineole 2-endo-monooxygenase / carbazole catabolic process / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen / iron ion binding /  heme binding heme bindingSimilarity search - Function | ||||||
| Biological species |   Citrobacter braakii (bacteria) Citrobacter braakii (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.554 Å MOLECULAR REPLACEMENT / Resolution: 1.554 Å | ||||||
 Authors Authors | Madrona, Y. / Tripathi, S.M. / Huiying, L. / Poulos, T.L. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2012 Journal: Biochemistry / Year: 2012Title: Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation. Authors: Madrona, Y. / Tripathi, S. / Li, H. / Poulos, T.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4fmx.cif.gz 4fmx.cif.gz | 482.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4fmx.ent.gz pdb4fmx.ent.gz | 401.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4fmx.json.gz 4fmx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fm/4fmx https://data.pdbj.org/pub/pdb/validation_reports/fm/4fmx ftp://data.pdbj.org/pub/pdb/validation_reports/fm/4fmx ftp://data.pdbj.org/pub/pdb/validation_reports/fm/4fmx | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4fb2C  4fyzC  4g3rC  1t2bS C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 2 molecules AB
| #1: Protein |  1,8-Cineole 2-endo-monooxygenase 1,8-Cineole 2-endo-monooxygenaseMass: 44787.879 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Citrobacter braakii (bacteria) / Gene: CIN A, cinA / Plasmid: pCWori-P450cin / Production host: Citrobacter braakii (bacteria) / Gene: CIN A, cinA / Plasmid: pCWori-P450cin / Production host:   Escherichia coli (E. coli) / Strain (production host): DH5-alpha / References: UniProt: Q8VQF6 Escherichia coli (E. coli) / Strain (production host): DH5-alpha / References: UniProt: Q8VQF6 |
|---|
-Non-polymers , 5 types, 885 molecules 
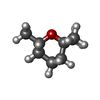







| #2: Chemical |  Heme B Heme B#3: Chemical |  Eucalyptol Eucalyptol#4: Chemical |  Sulfate Sulfate#5: Chemical |  Glycerol Glycerol#6: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.71 Å3/Da / Density % sol: 54.56 % |
|---|---|
Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 6.2 Details: 6-9% PEG 3350, 100mM BisTris pH 6.2, 100mM Lithium Sulfate, 5mM Cineole, VAPOR DIFFUSION, SITTING DROP, temperature 298K |
-Data collection
| Diffraction | Mean temperature: 70 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SSRL SSRL  / Beamline: BL9-2 / Wavelength: 0.98 Å / Beamline: BL9-2 / Wavelength: 0.98 Å |
| Detector | Type: MARMOSAIC 325 mm CCD / Detector: CCD / Date: Feb 28, 2010 / Details: mirrors |
| Radiation | Monochromator: GRAPHITE / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.98 Å / Relative weight: 1 : 0.98 Å / Relative weight: 1 |
| Reflection | Resolution: 1.55→50 Å / Num. all: 136152 / Num. obs: 136152 / % possible obs: 99.2 % / Observed criterion σ(F): 0.02 / Observed criterion σ(I): -3 / Redundancy: 3.3 % / Biso Wilson estimate: 17.1 Å2 / Rsym value: 0.074 / Net I/σ(I): 20.1 |
| Reflection shell | Resolution: 1.55→1.58 Å / Redundancy: 3.3 % / Mean I/σ(I) obs: 2.11 / Num. unique all: 6791 / Rsym value: 0.583 / % possible all: 99.7 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1T2B Resolution: 1.554→34.995 Å / SU ML: 0.23 / Cross valid method: THROUGHOUT / σ(F): 1.34 / Phase error: 21.47 / Stereochemistry target values: ML Details: In the B subunit, the carboxyl group of Asp227 comes within covalent bonding distance of the hydroxyl group in Tyr101. The 2mFo-DFc map is continuous in this region. This observation is ...Details: In the B subunit, the carboxyl group of Asp227 comes within covalent bonding distance of the hydroxyl group in Tyr101. The 2mFo-DFc map is continuous in this region. This observation is likely due to an X-ray induced structural change and is not present in the A subunit
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.73 Å / VDW probe radii: 1 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 49.24 Å2 / ksol: 0.382 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.554→34.995 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Refine-ID: X-RAY DIFFRACTION
|
 Movie
Movie Controller
Controller












 PDBj
PDBj




