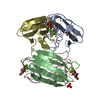
Entry Database : PDB / ID : 3zdvTitle Crystal structure of the LecB lectin from Pseudomonas aeruginosa in complex with Methyl 6-(2,4,6-trimethylphenylsulfonylamido)-6-deoxy-alpha-D-mannopyranoside FUCOSE-BINDING LECTIN PA-IIL Keywords / / Function / homology Function Domain/homology Component
/ / / / / / / Biological species PSEUDOMONAS AERUGINOSA (bacteria)Method / / / Resolution : 1.41 Å Authors Hauck, D. / Joachim, I. / Frommeyer, B. / Varrot, A. / Philipp, B. / MOller, H.M. / Imberty, A. / Exner, T.E. / Titz, A. Journal : Acs Chem.Biol. / Year : 2013Title : Discovery of Two Classes of Potent Glycomimetic Inhibitors of Pseudomonas Aeruginosa Lecb with Distinct Binding Modes.Authors : Hauck, D. / Joachim, I. / Frommeyer, B. / Varrot, A. / Philipp, B. / Moller, H.M. / Imberty, A. / Exner, T.E. / Titz, A. History Deposition Nov 30, 2012 Deposition site / Processing site Revision 1.0 Sep 18, 2013 Provider / Type Revision 1.1 Nov 13, 2013 Group Revision 1.2 Sep 13, 2017 Group / Structure summary / Category / struct / Item / _struct.titleRevision 1.3 Jul 29, 2020 Group Data collection / Derived calculations ... Data collection / Derived calculations / Other / Structure summary Category chem_comp / entity ... chem_comp / entity / pdbx_chem_comp_identifier / pdbx_database_status / pdbx_entity_nonpoly / pdbx_struct_conn_angle / struct_conn / struct_conn_type / struct_site / struct_site_gen Item _chem_comp.mon_nstd_flag / _chem_comp.name ... _chem_comp.mon_nstd_flag / _chem_comp.name / _chem_comp.type / _entity.pdbx_description / _pdbx_database_status.status_code_sf / _pdbx_entity_nonpoly.name / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr2_auth_seq_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.value / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn_type.id Description / Provider / Type Revision 1.4 Dec 20, 2023 Group Data collection / Database references ... Data collection / Database references / Refinement description / Structure summary Category chem_comp / chem_comp_atom ... chem_comp / chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model Item / _database_2.pdbx_DOI / _database_2.pdbx_database_accession
Show all Show less Remark 700 SHEET DETERMINATION METHOD: AUTHOR PROVIDED.
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords GLYCOMIMETIC /
GLYCOMIMETIC /  INHIBITOR
INHIBITOR Function and homology information
Function and homology information carbohydrate binding /
carbohydrate binding /  metal ion binding
metal ion binding
 PSEUDOMONAS AERUGINOSA (bacteria)
PSEUDOMONAS AERUGINOSA (bacteria) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.41 Å
MOLECULAR REPLACEMENT / Resolution: 1.41 Å  Authors
Authors Citation
Citation Journal: Acs Chem.Biol. / Year: 2013
Journal: Acs Chem.Biol. / Year: 2013 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3zdv.cif.gz
3zdv.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3zdv.ent.gz
pdb3zdv.ent.gz PDB format
PDB format 3zdv.json.gz
3zdv.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/zd/3zdv
https://data.pdbj.org/pub/pdb/validation_reports/zd/3zdv ftp://data.pdbj.org/pub/pdb/validation_reports/zd/3zdv
ftp://data.pdbj.org/pub/pdb/validation_reports/zd/3zdv
 Links
Links Assembly
Assembly
 Components
Components


 PSEUDOMONAS AERUGINOSA (bacteria) / Strain: PAO1 / Plasmid: PET25PA2 / Production host:
PSEUDOMONAS AERUGINOSA (bacteria) / Strain: PAO1 / Plasmid: PET25PA2 / Production host: 
 ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q9HYN5
ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q9HYN5 Methylglucoside
Methylglucoside









 Glycerol
Glycerol Ethylene glycol
Ethylene glycol Sulfate
Sulfate Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  SOLEIL
SOLEIL  / Beamline: PROXIMA 1 / Wavelength: 0.97857
/ Beamline: PROXIMA 1 / Wavelength: 0.97857  : 0.97857 Å / Relative weight: 1
: 0.97857 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller











 PDBj
PDBj




