[English] 日本語
 Yorodumi
Yorodumi- PDB-3oe6: Crystal structure of the CXCR4 chemokine receptor in complex with... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3oe6 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in I222 spacegroup | ||||||
 Components Components | C-X-C chemokine receptor type 4, Lysozyme Chimera | ||||||
 Keywords Keywords |  SIGNALING PROTEIN / SIGNALING PROTEIN /  HYDROLASE / HYDROLASE /  Structural Genomics / PSI-2 / Structural Genomics / PSI-2 /  Protein Structure Initiative / Accelerated Technologies Center for Gene to 3D Structure / ATCG3D / 7TM / Protein Structure Initiative / Accelerated Technologies Center for Gene to 3D Structure / ATCG3D / 7TM /  G protein-coupled receptor / G protein-coupled receptor /  GPCR / GPCR /  Signal transduction / Signal transduction /  Cancer / HIV-1 co-receptor / Cancer / HIV-1 co-receptor /  Chemokine / Chemokine /  CXCL12 / CXCL12 /  SDF1 / SDF1 /  isothiourea / IT1t / isothiourea / IT1t /  Chimera / T4L Fusion / Chimera / T4L Fusion /  Membrane protein / Membrane protein /  Transmembrane / SINGNALING PROTEIN / PSI-Biology / GPCR Network Transmembrane / SINGNALING PROTEIN / PSI-Biology / GPCR Network | ||||||
| Function / homology |  Function and homology information Function and homology informationC-X-C motif chemokine 12 receptor activity / regulation of viral process / positive regulation of vascular wound healing / positive regulation of macrophage migration inhibitory factor signaling pathway / positive regulation of mesenchymal stem cell migration / neuron recognition / response to ultrasound / response to tacrolimus / telencephalon cell migration / C-X-C chemokine receptor activity ...C-X-C motif chemokine 12 receptor activity / regulation of viral process / positive regulation of vascular wound healing / positive regulation of macrophage migration inhibitory factor signaling pathway / positive regulation of mesenchymal stem cell migration / neuron recognition / response to ultrasound / response to tacrolimus / telencephalon cell migration / C-X-C chemokine receptor activity / Specification of primordial germ cells / CXCL12-activated CXCR4 signaling pathway /  myosin light chain binding / myosin light chain binding /  myelin maintenance / positive regulation of vasculature development / myelin maintenance / positive regulation of vasculature development /  regulation of programmed cell death / C-C chemokine receptor activity / endothelial tube morphogenesis / endothelial cell differentiation / C-C chemokine binding / Signaling by ROBO receptors / regulation of programmed cell death / C-C chemokine receptor activity / endothelial tube morphogenesis / endothelial cell differentiation / C-C chemokine binding / Signaling by ROBO receptors /  regulation of chemotaxis / cellular response to organonitrogen compound / positive regulation of chemotaxis / Formation of definitive endoderm / positive regulation of dendrite extension / anchoring junction / Chemokine receptors bind chemokines / dendritic cell chemotaxis / positive regulation of oligodendrocyte differentiation / epithelial cell development / cell leading edge / cellular response to cytokine stimulus / regulation of chemotaxis / cellular response to organonitrogen compound / positive regulation of chemotaxis / Formation of definitive endoderm / positive regulation of dendrite extension / anchoring junction / Chemokine receptors bind chemokines / dendritic cell chemotaxis / positive regulation of oligodendrocyte differentiation / epithelial cell development / cell leading edge / cellular response to cytokine stimulus /  small molecule binding / detection of temperature stimulus involved in sensory perception of pain / regulation of calcium ion transport / Binding and entry of HIV virion / small molecule binding / detection of temperature stimulus involved in sensory perception of pain / regulation of calcium ion transport / Binding and entry of HIV virion /  coreceptor activity / detection of mechanical stimulus involved in sensory perception of pain / coreceptor activity / detection of mechanical stimulus involved in sensory perception of pain /  regulation of cell adhesion / cardiac muscle contraction / viral release from host cell by cytolysis / regulation of cell adhesion / cardiac muscle contraction / viral release from host cell by cytolysis /  neurogenesis / cell chemotaxis / peptidoglycan catabolic process / response to activity / neurogenesis / cell chemotaxis / peptidoglycan catabolic process / response to activity /  ubiquitin binding / G protein-coupled receptor activity / calcium-mediated signaling / ubiquitin binding / G protein-coupled receptor activity / calcium-mediated signaling /  brain development / brain development /  neuron migration / response to virus / cell wall macromolecule catabolic process / cellular response to xenobiotic stimulus / late endosome / virus receptor activity / neuron migration / response to virus / cell wall macromolecule catabolic process / cellular response to xenobiotic stimulus / late endosome / virus receptor activity /  lysozyme / lysozyme /  lysozyme activity / positive regulation of cold-induced thermogenesis / lysozyme activity / positive regulation of cold-induced thermogenesis /  actin binding / positive regulation of cytosolic calcium ion concentration / G alpha (i) signalling events / cytoplasmic vesicle / host cell cytoplasm / actin binding / positive regulation of cytosolic calcium ion concentration / G alpha (i) signalling events / cytoplasmic vesicle / host cell cytoplasm /  lysosome / response to hypoxia / lysosome / response to hypoxia /  early endosome / positive regulation of cell migration / defense response to bacterium / early endosome / positive regulation of cell migration / defense response to bacterium /  immune response / immune response /  inflammatory response / G protein-coupled receptor signaling pathway / external side of plasma membrane / apoptotic process / inflammatory response / G protein-coupled receptor signaling pathway / external side of plasma membrane / apoptotic process /  ubiquitin protein ligase binding / ubiquitin protein ligase binding /  cell surface / protein-containing complex / extracellular exosome / cell surface / protein-containing complex / extracellular exosome /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo Sapiens (human) Homo Sapiens (human)  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.2 Å MOLECULAR REPLACEMENT / Resolution: 3.2 Å | ||||||
 Authors Authors | Wu, B. / Mol, C.D. / Han, G.W. / Katritch, V. / Chien, E.Y.T. / Liu, W. / Cherezov, V. / Stevens, R.C. / Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) / GPCR Network (GPCR) | ||||||
 Citation Citation |  Journal: Science / Year: 2010 Journal: Science / Year: 2010Title: Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Authors: Wu, B. / Chien, E.Y. / Mol, C.D. / Fenalti, G. / Liu, W. / Katritch, V. / Abagyan, R. / Brooun, A. / Wells, P. / Bi, F.C. / Hamel, D.J. / Kuhn, P. / Handel, T.M. / Cherezov, V. / Stevens, R.C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3oe6.cif.gz 3oe6.cif.gz | 183.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3oe6.ent.gz pdb3oe6.ent.gz | 151.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3oe6.json.gz 3oe6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/oe/3oe6 https://data.pdbj.org/pub/pdb/validation_reports/oe/3oe6 ftp://data.pdbj.org/pub/pdb/validation_reports/oe/3oe6 ftp://data.pdbj.org/pub/pdb/validation_reports/oe/3oe6 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3oduC  3oe0C  3oe8C  3oe9C C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 57537.426 Da / Num. of mol.: 1 Fragment: CXCR4 residues 2-228, LYSOZYME residues 1002-1161, CXCR4 residues 231-325 Mutation: L125W, C1054T, C1097T Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo Sapiens (human), (gene. exp.) Homo Sapiens (human), (gene. exp.)   Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus)Gene: CXCR4, CXCR4_HUMAN,E / Plasmid: pFastBac / Production host:   Spodoptera frugiperda (fall armyworm) / Strain (production host): Sf9 / References: UniProt: P61073, UniProt: P00720, Spodoptera frugiperda (fall armyworm) / Strain (production host): Sf9 / References: UniProt: P61073, UniProt: P00720,  lysozyme lysozyme | ||||
|---|---|---|---|---|---|
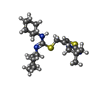
| #2: Chemical | ChemComp-ITD / ( | ||||
| #3: Chemical | | #4: Chemical | ChemComp-GOL / |  Glycerol GlycerolSequence details | THE PROTEIN IS A FUSION PROTEIN WITH RESIDUES ASN1002-TYR1161 OF T4 LYSOZYME INSERTED BETWEEN ...THE PROTEIN IS A FUSION PROTEIN WITH RESIDUES ASN1002-TYR1161 OF T4 LYSOZYME INSERTED BETWEEN SER229 AND LYS230 OF CXCR4, AS INDICATED AS CXCR4-1 IN THE PUBLICATIO | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 4 X-RAY DIFFRACTION / Number of used crystals: 4 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.93 Å3/Da / Density % sol: 57.96 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: lipidic cubic phase / pH: 5.5 Details: Lipidic cubic phase made of monoolein and cholesterol, 20-26% PEG400, 0.3M Sodium malonate, 5mM Nickel chloride, 0.1M Sodium citrate pH 5.0-5.5, LIPIDIC CUBIC PHASE, temperature 293K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  APS APS  / Beamline: 23-ID-D / Wavelength: 1.033 Å / Beamline: 23-ID-D / Wavelength: 1.033 Å |
| Detector | Type: MARMOSAIC 300 mm CCD / Detector: CCD / Date: Feb 25, 2010 / Details: mirrors |
| Radiation | Monochromator: DOUBLE CRYSTAL / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.033 Å / Relative weight: 1 : 1.033 Å / Relative weight: 1 |
| Reflection | Resolution: 3.2→50 Å / Num. obs: 10233 / % possible obs: 94.4 % / Redundancy: 3.6 % / Biso Wilson estimate: 89.11 Å2 / Rmerge(I) obs: 0.136 / Net I/σ(I): 10.2 |
| Reflection shell | Resolution: 3.3→3.42 Å / Redundancy: 2.2 % / Rmerge(I) obs: 0.67 / Mean I/σ(I) obs: 1.5 / % possible all: 78.2 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT / Cor.coef. Fo:Fc: 0.8474 / Cor.coef. Fo:Fc free: 0.7467 / Highest resolution: 3.2 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber MOLECULAR REPLACEMENT / Cor.coef. Fo:Fc: 0.8474 / Cor.coef. Fo:Fc free: 0.7467 / Highest resolution: 3.2 Å / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 69.12 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze | Luzzati coordinate error obs: 0.578 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 3.2 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.2→3.58 Å / Total num. of bins used: 5
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj