+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3iar | ||||||
|---|---|---|---|---|---|---|---|
| Title | The crystal structure of human adenosine deaminase | ||||||
 Components Components | Adenosine deaminase | ||||||
 Keywords Keywords |  HYDROLASE / HYDROLASE /  deaminase / deaminase /  adenosine deaminase / adenosine deaminase /  adenosine / adenosine /  purine metabolism / purine metabolism /  Structural Genomics / Structural Genomics /  Structural Genomics Consortium / SGC / Disease mutation / Hereditary hemolytic anemia / Structural Genomics Consortium / SGC / Disease mutation / Hereditary hemolytic anemia /  Nucleotide metabolism / SCID Nucleotide metabolism / SCID | ||||||
| Function / homology |  Function and homology information Function and homology informationpurine nucleotide salvage / Defective ADA disrupts (deoxy)adenosine deamination / mature B cell apoptotic process / xanthine biosynthetic process / negative regulation of penile erection / negative regulation of mucus secretion /  penile erection / positive regulation of germinal center formation / negative regulation of adenosine receptor signaling pathway / inosine biosynthetic process ...purine nucleotide salvage / Defective ADA disrupts (deoxy)adenosine deamination / mature B cell apoptotic process / xanthine biosynthetic process / negative regulation of penile erection / negative regulation of mucus secretion / penile erection / positive regulation of germinal center formation / negative regulation of adenosine receptor signaling pathway / inosine biosynthetic process ...purine nucleotide salvage / Defective ADA disrupts (deoxy)adenosine deamination / mature B cell apoptotic process / xanthine biosynthetic process / negative regulation of penile erection / negative regulation of mucus secretion /  penile erection / positive regulation of germinal center formation / negative regulation of adenosine receptor signaling pathway / inosine biosynthetic process / cytoplasmic vesicle lumen / 2'-deoxyadenosine deaminase activity / amide catabolic process / penile erection / positive regulation of germinal center formation / negative regulation of adenosine receptor signaling pathway / inosine biosynthetic process / cytoplasmic vesicle lumen / 2'-deoxyadenosine deaminase activity / amide catabolic process /  adenosine deaminase / germinal center B cell differentiation / adenosine catabolic process / purine-containing compound salvage / adenosine deaminase / germinal center B cell differentiation / adenosine catabolic process / purine-containing compound salvage /  deaminase activity / deaminase activity /  adenosine deaminase activity / hypoxanthine salvage / deoxyadenosine catabolic process / dAMP catabolic process / adenosine metabolic process / AMP catabolic process / positive regulation of T cell differentiation in thymus / dATP catabolic process / negative regulation of leukocyte migration / mucus secretion / Ribavirin ADME / regulation of cell-cell adhesion mediated by integrin / response to purine-containing compound / embryonic digestive tract development / allantoin metabolic process / trophectodermal cell differentiation / GMP salvage / Purine salvage / positive regulation of smooth muscle contraction / Peyer's patch development / germinal center formation / negative regulation of mature B cell apoptotic process / AMP salvage / negative regulation of thymocyte apoptotic process / anchoring junction / positive regulation of alpha-beta T cell differentiation / alpha-beta T cell differentiation / positive regulation of heart rate / leukocyte migration / lung alveolus development / positive regulation of T cell receptor signaling pathway / thymocyte apoptotic process / B cell proliferation / smooth muscle contraction / : / positive regulation of calcium-mediated signaling / positive regulation of B cell proliferation / adenosine deaminase activity / hypoxanthine salvage / deoxyadenosine catabolic process / dAMP catabolic process / adenosine metabolic process / AMP catabolic process / positive regulation of T cell differentiation in thymus / dATP catabolic process / negative regulation of leukocyte migration / mucus secretion / Ribavirin ADME / regulation of cell-cell adhesion mediated by integrin / response to purine-containing compound / embryonic digestive tract development / allantoin metabolic process / trophectodermal cell differentiation / GMP salvage / Purine salvage / positive regulation of smooth muscle contraction / Peyer's patch development / germinal center formation / negative regulation of mature B cell apoptotic process / AMP salvage / negative regulation of thymocyte apoptotic process / anchoring junction / positive regulation of alpha-beta T cell differentiation / alpha-beta T cell differentiation / positive regulation of heart rate / leukocyte migration / lung alveolus development / positive regulation of T cell receptor signaling pathway / thymocyte apoptotic process / B cell proliferation / smooth muscle contraction / : / positive regulation of calcium-mediated signaling / positive regulation of B cell proliferation /  T cell activation / xenobiotic metabolic process / liver development / calcium-mediated signaling / placenta development / negative regulation of inflammatory response / T cell differentiation in thymus / T cell receptor signaling pathway / T cell activation / xenobiotic metabolic process / liver development / calcium-mediated signaling / placenta development / negative regulation of inflammatory response / T cell differentiation in thymus / T cell receptor signaling pathway /  lysosome / response to hypoxia / lysosome / response to hypoxia /  cell adhesion / external side of plasma membrane / cell adhesion / external side of plasma membrane /  cell surface / zinc ion binding / cell surface / zinc ion binding /  membrane / membrane /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 1.52 Å MOLECULAR REPLACEMENT / Resolution: 1.52 Å | ||||||
 Authors Authors | Ugochukwu, E. / Zhang, Y. / Hapka, E. / Yue, W.W. / Bray, J.E. / Muniz, J. / Burgess-Brown, N. / Chaikuad, A. / von Delft, F. / Bountra, C. ...Ugochukwu, E. / Zhang, Y. / Hapka, E. / Yue, W.W. / Bray, J.E. / Muniz, J. / Burgess-Brown, N. / Chaikuad, A. / von Delft, F. / Bountra, C. / Arrowsmith, C.H. / Weigelt, J. / Edwards, A. / Kavanagh, K.L. / Oppermann, U. / Structural Genomics Consortium (SGC) | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: The crystal structure of human adenosine deaminase Authors: Ugochukwu, E. / Zhang, Y. / Hapka, E. / Yue, W.W. / Bray, J.E. / Muniz, J. / Burgess-Brown, N. / Chaikuad, A. / Kavanagh, K.L. / Oppermann, U. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3iar.cif.gz 3iar.cif.gz | 172.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3iar.ent.gz pdb3iar.ent.gz | 132.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3iar.json.gz 3iar.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ia/3iar https://data.pdbj.org/pub/pdb/validation_reports/ia/3iar ftp://data.pdbj.org/pub/pdb/validation_reports/ia/3iar ftp://data.pdbj.org/pub/pdb/validation_reports/ia/3iar | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1krmS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein |  / Adenosine aminohydrolase / Adenosine aminohydrolaseMass: 41381.988 Da / Num. of mol.: 1 / Fragment: UNP residues 5-363 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: ADA / Plasmid: pNIC-CTHF / Production host: Homo sapiens (human) / Gene: ADA / Plasmid: pNIC-CTHF / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21(DE3)-R3-pRARE2 / References: UniProt: P00813, Escherichia coli (E. coli) / Strain (production host): BL21(DE3)-R3-pRARE2 / References: UniProt: P00813,  adenosine deaminase adenosine deaminase |
|---|
-Non-polymers , 5 types, 696 molecules 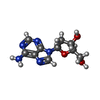








| #2: Chemical | ChemComp-3D1 / ( Deoxyadenosine Deoxyadenosine | ||||
|---|---|---|---|---|---|
| #3: Chemical | ChemComp-NI /  Nickel Nickel | ||||
| #4: Chemical |  Nitrate Nitrate#5: Chemical | ChemComp-GOL / |  Glycerol Glycerol#6: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.08 Å3/Da / Density % sol: 40.84 % |
|---|---|
Crystal grow | Temperature: 277 K / Method: vapor diffusion, sitting drop Details: 20% PEG 3350, 0.20M NaNO3, VAPOR DIFFUSION, SITTING DROP, temperature 277K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU FR-E SUPERBRIGHT / Wavelength: 1.5418 Å ROTATING ANODE / Type: RIGAKU FR-E SUPERBRIGHT / Wavelength: 1.5418 Å |
| Detector | Type: RIGAKU RAXIS IV / Detector: IMAGE PLATE / Date: Jul 1, 2009 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 1.52→29.698 Å / Num. obs: 52538 / % possible obs: 97.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 4.2 % / Rmerge(I) obs: 0.048 / Rsym value: 0.048 / Net I/σ(I): 18.6 |
| Reflection shell | Resolution: 1.52→1.6 Å / Redundancy: 2.2 % / Rmerge(I) obs: 0.251 / Mean I/σ(I) obs: 3.1 / Num. unique all: 14424 / Rsym value: 0.251 / % possible all: 86.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB entry 1KRM Resolution: 1.52→29.698 Å / SU ML: 0.55 / Cross valid method: THROUGHOUT / σ(F): 1.35 / σ(I): 0 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 32.941 Å2 / ksol: 0.318 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.52→29.698 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj






