Entry Database : PDB / ID : 3i5sTitle Crystal structure of PI3K SH3 Phosphatidylinositol 3-kinase regulatory subunit alpha Keywords / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 3 Å Authors Batra-Safferling, R. / Granzin, J. / Modder, S. / Hoffmann, S. / Willbold, D. Journal : Biol.Chem. / Year : 2010Title : Structural studies of the phosphatidylinositol 3-kinase (PI3K) SH3 domain in complex with a peptide ligand: role of the anchor residue in ligand binding.Authors : Batra-Safferling, R. / Granzin, J. / Modder, S. / Hoffmann, S. / Willbold, D. History Deposition Jul 6, 2009 Deposition site / Processing site Revision 1.0 Mar 2, 2010 Provider / Type Revision 1.1 Jul 13, 2011 Group Revision 1.2 Sep 6, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_ncs_dom_lim / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ncs_dom_lim.beg_auth_comp_id / _struct_ncs_dom_lim.beg_label_asym_id / _struct_ncs_dom_lim.beg_label_comp_id / _struct_ncs_dom_lim.beg_label_seq_id / _struct_ncs_dom_lim.end_auth_comp_id / _struct_ncs_dom_lim.end_label_asym_id / _struct_ncs_dom_lim.end_label_comp_id / _struct_ncs_dom_lim.end_label_seq_id / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords PROTEIN BINDING /
PROTEIN BINDING /  SH3 domain /
SH3 domain /  Alternative splicing / Disease mutation / Host-virus interaction /
Alternative splicing / Disease mutation / Host-virus interaction /  Phosphoprotein / Polymorphism /
Phosphoprotein / Polymorphism /  SH2 domain / Ubl conjugation
SH2 domain / Ubl conjugation Function and homology information
Function and homology information transmembrane receptor protein tyrosine kinase adaptor activity / RHOD GTPase cycle / Signaling by cytosolic FGFR1 fusion mutants / positive regulation of endoplasmic reticulum unfolded protein response / enzyme-substrate adaptor activity / phosphatidylinositol 3-kinase complex, class IA /
transmembrane receptor protein tyrosine kinase adaptor activity / RHOD GTPase cycle / Signaling by cytosolic FGFR1 fusion mutants / positive regulation of endoplasmic reticulum unfolded protein response / enzyme-substrate adaptor activity / phosphatidylinositol 3-kinase complex, class IA /  phosphatidylinositol 3-kinase complex / Nephrin family interactions / RND1 GTPase cycle / Costimulation by the CD28 family / RND2 GTPase cycle / MET activates PI3K/AKT signaling / PI3K/AKT activation / RND3 GTPase cycle / positive regulation of leukocyte migration / positive regulation of filopodium assembly / negative regulation of stress fiber assembly / growth hormone receptor signaling pathway /
phosphatidylinositol 3-kinase complex / Nephrin family interactions / RND1 GTPase cycle / Costimulation by the CD28 family / RND2 GTPase cycle / MET activates PI3K/AKT signaling / PI3K/AKT activation / RND3 GTPase cycle / positive regulation of leukocyte migration / positive regulation of filopodium assembly / negative regulation of stress fiber assembly / growth hormone receptor signaling pathway /  insulin binding / RHOV GTPase cycle / RHOB GTPase cycle / negative regulation of cell-matrix adhesion / Signaling by ALK / GP1b-IX-V activation signalling / PI-3K cascade:FGFR3 / Erythropoietin activates Phosphoinositide-3-kinase (PI3K) / PI-3K cascade:FGFR2 / RHOJ GTPase cycle / PI-3K cascade:FGFR4 / RHOC GTPase cycle / PI-3K cascade:FGFR1 / negative regulation of osteoclast differentiation / intracellular glucose homeostasis / phosphatidylinositol phosphate biosynthetic process / CD28 dependent PI3K/Akt signaling / Synthesis of PIPs at the plasma membrane / CDC42 GTPase cycle / RHOU GTPase cycle / PI3K events in ERBB2 signaling / RHOG GTPase cycle / T cell differentiation / RET signaling / extrinsic apoptotic signaling pathway via death domain receptors /
insulin binding / RHOV GTPase cycle / RHOB GTPase cycle / negative regulation of cell-matrix adhesion / Signaling by ALK / GP1b-IX-V activation signalling / PI-3K cascade:FGFR3 / Erythropoietin activates Phosphoinositide-3-kinase (PI3K) / PI-3K cascade:FGFR2 / RHOJ GTPase cycle / PI-3K cascade:FGFR4 / RHOC GTPase cycle / PI-3K cascade:FGFR1 / negative regulation of osteoclast differentiation / intracellular glucose homeostasis / phosphatidylinositol phosphate biosynthetic process / CD28 dependent PI3K/Akt signaling / Synthesis of PIPs at the plasma membrane / CDC42 GTPase cycle / RHOU GTPase cycle / PI3K events in ERBB2 signaling / RHOG GTPase cycle / T cell differentiation / RET signaling / extrinsic apoptotic signaling pathway via death domain receptors /  insulin receptor substrate binding /
insulin receptor substrate binding /  Interleukin-3, Interleukin-5 and GM-CSF signaling / PI3K Cascade / RHOA GTPase cycle / RAC2 GTPase cycle / RAC3 GTPase cycle / Role of phospholipids in phagocytosis / GAB1 signalosome / Role of LAT2/NTAL/LAB on calcium mobilization / Interleukin receptor SHC signaling / positive regulation of lamellipodium assembly /
Interleukin-3, Interleukin-5 and GM-CSF signaling / PI3K Cascade / RHOA GTPase cycle / RAC2 GTPase cycle / RAC3 GTPase cycle / Role of phospholipids in phagocytosis / GAB1 signalosome / Role of LAT2/NTAL/LAB on calcium mobilization / Interleukin receptor SHC signaling / positive regulation of lamellipodium assembly /  phosphatidylinositol 3-kinase binding / Signaling by PDGFRA transmembrane, juxtamembrane and kinase domain mutants / Signaling by PDGFRA extracellular domain mutants / Signaling by FGFR4 in disease / Signaling by FLT3 ITD and TKD mutants / Signaling by FGFR3 in disease / GPVI-mediated activation cascade / Tie2 Signaling / Signaling by FGFR2 in disease / RAC1 GTPase cycle /
phosphatidylinositol 3-kinase binding / Signaling by PDGFRA transmembrane, juxtamembrane and kinase domain mutants / Signaling by PDGFRA extracellular domain mutants / Signaling by FGFR4 in disease / Signaling by FLT3 ITD and TKD mutants / Signaling by FGFR3 in disease / GPVI-mediated activation cascade / Tie2 Signaling / Signaling by FGFR2 in disease / RAC1 GTPase cycle /  insulin-like growth factor receptor binding / Signaling by FLT3 fusion proteins / FLT3 Signaling / Signaling by FGFR1 in disease / Interleukin-7 signaling / phosphotyrosine residue binding / response to endoplasmic reticulum stress / Downstream signal transduction / substrate adhesion-dependent cell spreading / B cell differentiation / osteoclast differentiation / positive regulation of RNA splicing / insulin-like growth factor receptor signaling pathway / Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants / phosphatidylinositol 3-kinase/protein kinase B signal transduction / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / Regulation of signaling by CBL
insulin-like growth factor receptor binding / Signaling by FLT3 fusion proteins / FLT3 Signaling / Signaling by FGFR1 in disease / Interleukin-7 signaling / phosphotyrosine residue binding / response to endoplasmic reticulum stress / Downstream signal transduction / substrate adhesion-dependent cell spreading / B cell differentiation / osteoclast differentiation / positive regulation of RNA splicing / insulin-like growth factor receptor signaling pathway / Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants / phosphatidylinositol 3-kinase/protein kinase B signal transduction / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / Regulation of signaling by CBL
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3 Å
MOLECULAR REPLACEMENT / Resolution: 3 Å  Authors
Authors Citation
Citation Journal: Biol.Chem. / Year: 2010
Journal: Biol.Chem. / Year: 2010 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3i5s.cif.gz
3i5s.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3i5s.ent.gz
pdb3i5s.ent.gz PDB format
PDB format 3i5s.json.gz
3i5s.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/i5/3i5s
https://data.pdbj.org/pub/pdb/validation_reports/i5/3i5s ftp://data.pdbj.org/pub/pdb/validation_reports/i5/3i5s
ftp://data.pdbj.org/pub/pdb/validation_reports/i5/3i5s

 Links
Links Assembly
Assembly




 Components
Components
 Homo sapiens (human) / Gene: GRB1, PIK3R1 / Plasmid: pGEX / Production host:
Homo sapiens (human) / Gene: GRB1, PIK3R1 / Plasmid: pGEX / Production host: 
 Escherichia coli (E. coli) / Strain (production host): BL21 / References: UniProt: P27986
Escherichia coli (E. coli) / Strain (production host): BL21 / References: UniProt: P27986 Sulfate
Sulfate X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  ESRF
ESRF  / Beamline: ID14-1 / Wavelength: 0.934 Å
/ Beamline: ID14-1 / Wavelength: 0.934 Å : 0.934 Å / Relative weight: 1
: 0.934 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller






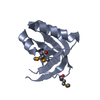


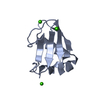



 PDBj
PDBj



















