[English] 日本語
 Yorodumi
Yorodumi- PDB-3edu: Crystal structure of the ankyrin-binding domain of human erythroi... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3edu | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of the ankyrin-binding domain of human erythroid spectrin | ||||||
 Components Components | Spectrin beta chain, erythrocyte | ||||||
 Keywords Keywords |  STRUCTURAL PROTEIN / STRUCTURAL PROTEIN /  spectrin / spectrin /  ankyrin / ankyrin-binding domain / Actin capping / Actin-binding / ankyrin / ankyrin-binding domain / Actin capping / Actin-binding /  Cytoskeleton / Disease mutation / Cytoskeleton / Disease mutation /  Elliptocytosis / Hereditary hemolytic anemia / Elliptocytosis / Hereditary hemolytic anemia /  Phosphoprotein Phosphoprotein | ||||||
| Function / homology |  Function and homology information Function and homology information spectrin / spectrin-associated cytoskeleton / modification of postsynaptic actin cytoskeleton / actin filament capping / Interaction between L1 and Ankyrins / spectrin / spectrin-associated cytoskeleton / modification of postsynaptic actin cytoskeleton / actin filament capping / Interaction between L1 and Ankyrins /  ankyrin binding / cortical actin cytoskeleton / COPI-mediated anterograde transport / NCAM signaling for neurite out-growth / cell projection ... ankyrin binding / cortical actin cytoskeleton / COPI-mediated anterograde transport / NCAM signaling for neurite out-growth / cell projection ... spectrin / spectrin-associated cytoskeleton / modification of postsynaptic actin cytoskeleton / actin filament capping / Interaction between L1 and Ankyrins / spectrin / spectrin-associated cytoskeleton / modification of postsynaptic actin cytoskeleton / actin filament capping / Interaction between L1 and Ankyrins /  ankyrin binding / cortical actin cytoskeleton / COPI-mediated anterograde transport / NCAM signaling for neurite out-growth / cell projection / cytoplasmic side of plasma membrane / structural constituent of cytoskeleton / ankyrin binding / cortical actin cytoskeleton / COPI-mediated anterograde transport / NCAM signaling for neurite out-growth / cell projection / cytoplasmic side of plasma membrane / structural constituent of cytoskeleton /  actin filament binding / actin filament binding /  actin cytoskeleton / actin cytoskeleton /  cell junction / cell junction /  actin binding / actin cytoskeleton organization / RAF/MAP kinase cascade / postsynapse / glutamatergic synapse / actin binding / actin cytoskeleton organization / RAF/MAP kinase cascade / postsynapse / glutamatergic synapse /  cell surface / protein-containing complex / cell surface / protein-containing complex /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  SAD / Resolution: 2.1 Å SAD / Resolution: 2.1 Å | ||||||
 Authors Authors | Simonovic, M. / Stabach, P. / Simonovic, I. / Steitz, T.A. / Morrow, J.S. | ||||||
 Citation Citation |  Journal: Blood / Year: 2009 Journal: Blood / Year: 2009Title: The structure of the ankyrin-binding site of {beta}-spectrin reveals how tandem spectrin-repeats generate unique ligand-binding properties Authors: Stabach, P.R. / Simonovic, I. / Ranieri, M.A. / Aboodi, M.S. / Steitz, T.A. / Simonovic, M. / Morrow, J.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3edu.cif.gz 3edu.cif.gz | 54.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3edu.ent.gz pdb3edu.ent.gz | 39.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3edu.json.gz 3edu.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ed/3edu https://data.pdbj.org/pub/pdb/validation_reports/ed/3edu ftp://data.pdbj.org/pub/pdb/validation_reports/ed/3edu ftp://data.pdbj.org/pub/pdb/validation_reports/ed/3edu | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 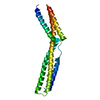
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 24810.543 Da / Num. of mol.: 1 / Fragment: Spectrin 14-Spectrin 15 di-repeat Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: SPTB, SPTB1 / Production host: Homo sapiens (human) / Gene: SPTB, SPTB1 / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21 (DE3) / References: UniProt: P11277 Escherichia coli (E. coli) / Strain (production host): BL21 (DE3) / References: UniProt: P11277 |
|---|---|
| #2: Water | ChemComp-HOH /  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.69 Å3/Da / Density % sol: 54.28 % |
|---|---|
Crystal grow | Temperature: 289 K / Method: vapor diffusion, sitting drop / pH: 6.2 Details: 0.1M bis-tris-propane, pH 6.2, 0.2M KSCN, 20% PEG 3,350, 3-10mM spermine, VAPOR DIFFUSION, SITTING DROP, temperature 289K |
-Data collection
| Diffraction |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| |||||||||||||||
| Detector |
| |||||||||||||||
| Radiation |
| |||||||||||||||
| Radiation wavelength | Wavelength : 0.9793 Å / Relative weight: 1 : 0.9793 Å / Relative weight: 1 | |||||||||||||||
| Reflection | Resolution: 2.1→31 Å / Num. all: 15823 / Num. obs: 15696 / % possible obs: 99.2 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 11.5 % / Biso Wilson estimate: 48.2 Å2 / Rsym value: 0.08 | |||||||||||||||
| Reflection shell | Resolution: 2.1→2.2 Å / Redundancy: 6.3 % / Mean I/σ(I) obs: 3 / Rsym value: 0.52 / % possible all: 96.3 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  SAD / Resolution: 2.1→31 Å / SU ML: 0.29 / σ(F): 1.34 / Stereochemistry target values: ML SAD / Resolution: 2.1→31 Å / SU ML: 0.29 / σ(F): 1.34 / Stereochemistry target values: ML
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.8 Å / VDW probe radii: 0.8 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 85.458 Å2 / ksol: 0.382 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 58.4 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→31 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj









