+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2vge | ||||||
|---|---|---|---|---|---|---|---|
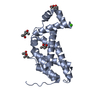
| Title | Crystal structure of the C-terminal region of human iASPP | ||||||
 Components Components | RELA-ASSOCIATED INHIBITOR | ||||||
 Keywords Keywords |  TRANSCRIPTION / IASPP / TRANSCRIPTION / IASPP /  NUCLEUS / NUCLEUS /  APOPTOSIS / APOPTOSIS /  REPRESSOR / REPRESSOR /  CYTOPLASM / CYTOPLASM /  PHOSPHORYLATION / P53 BINDING PROTEIN / ANK REPEAT / PHOSPHORYLATION / P53 BINDING PROTEIN / ANK REPEAT /  SH3 DOMAIN / ANKYRIN REPEATS / SH3 DOMAIN / ANKYRIN REPEATS /  ALTERNATIVE SPLICING / ALTERNATIVE SPLICING /  TRANSCRIPTION REGULATION TRANSCRIPTION REGULATION | ||||||
| Function / homology |  Function and homology information Function and homology informationmulticellular organismal-level homeostasis / cardiac right ventricle morphogenesis / embryonic camera-type eye development /  hair cycle / ventricular cardiac muscle tissue development / Regulation of TP53 Activity through Association with Co-factors / hair cycle / ventricular cardiac muscle tissue development / Regulation of TP53 Activity through Association with Co-factors /  intercellular bridge / cardiac muscle contraction / post-embryonic development / positive regulation of cell differentiation ...multicellular organismal-level homeostasis / cardiac right ventricle morphogenesis / embryonic camera-type eye development / intercellular bridge / cardiac muscle contraction / post-embryonic development / positive regulation of cell differentiation ...multicellular organismal-level homeostasis / cardiac right ventricle morphogenesis / embryonic camera-type eye development /  hair cycle / ventricular cardiac muscle tissue development / Regulation of TP53 Activity through Association with Co-factors / hair cycle / ventricular cardiac muscle tissue development / Regulation of TP53 Activity through Association with Co-factors /  intercellular bridge / cardiac muscle contraction / post-embryonic development / positive regulation of cell differentiation / multicellular organism growth / transcription corepressor activity / intercellular bridge / cardiac muscle contraction / post-embryonic development / positive regulation of cell differentiation / multicellular organism growth / transcription corepressor activity /  cell junction / cell junction /  cadherin binding / apoptotic process / regulation of transcription by RNA polymerase II / negative regulation of transcription by RNA polymerase II / cadherin binding / apoptotic process / regulation of transcription by RNA polymerase II / negative regulation of transcription by RNA polymerase II /  nucleoplasm / identical protein binding / nucleoplasm / identical protein binding /  nucleus / nucleus /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   HOMO SAPIENS (human) HOMO SAPIENS (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.1 Å MOLECULAR REPLACEMENT / Resolution: 2.1 Å | ||||||
 Authors Authors | Robinson, R.A. / Lu, X. / Jones, E.Y. / Siebold, C. | ||||||
 Citation Citation |  Journal: Structure / Year: 2008 Journal: Structure / Year: 2008Title: Biochemical and Structural Studies of Aspp Proteins Reveal Differential Binding to P53, P63 and P73 Authors: Robinson, R.A. / Lu, X. / Jones, E.Y. / Siebold, C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2vge.cif.gz 2vge.cif.gz | 56.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2vge.ent.gz pdb2vge.ent.gz | 39.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2vge.json.gz 2vge.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/vg/2vge https://data.pdbj.org/pub/pdb/validation_reports/vg/2vge ftp://data.pdbj.org/pub/pdb/validation_reports/vg/2vge ftp://data.pdbj.org/pub/pdb/validation_reports/vg/2vge | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1ycsS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 25314.373 Da / Num. of mol.: 1 / Fragment: P53 (CORE) BINDING DOMAIN, RESIDUES 607-828 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   HOMO SAPIENS (human) / Plasmid: PET 22B / Production host: HOMO SAPIENS (human) / Plasmid: PET 22B / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): ROSETTA (DE3) PLYSS / References: UniProt: Q8WUF5 ESCHERICHIA COLI (E. coli) / Strain (production host): ROSETTA (DE3) PLYSS / References: UniProt: Q8WUF5 |
|---|---|
| #2: Water | ChemComp-HOH /  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.09 Å3/Da / Density % sol: 40.73 % / Description: NONE |
|---|---|
Crystal grow | Details: 22% PEG3350, 0.3 M KCL2, 17.5% MPD |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID14-2 / Wavelength: 0.97566 / Beamline: ID14-2 / Wavelength: 0.97566 |
| Detector | Type: ADSC CCD / Detector: CCD / Date: Dec 15, 2003 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.97566 Å / Relative weight: 1 : 0.97566 Å / Relative weight: 1 |
| Reflection | Resolution: 2.1→30 Å / Num. obs: 12454 / % possible obs: 99.1 % / Observed criterion σ(I): 0 / Redundancy: 4.3 % / Biso Wilson estimate: 15.1 Å2 / Rmerge(I) obs: 0.12 / Net I/σ(I): 10.3 |
| Reflection shell | Resolution: 2.1→2.2 Å / Redundancy: 2.7 % / Rmerge(I) obs: 0.72 / Mean I/σ(I) obs: 1.9 / % possible all: 96 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1YCS Resolution: 2.1→29.43 Å / Rfactor Rfree error: 0.011 / Data cutoff high absF: 219304.74 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 46.9959 Å2 / ksol: 0.361446 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 32.8 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.1→29.43 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.1→2.23 Å / Rfactor Rfree error: 0.042 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj






