+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2rup | ||||||
|---|---|---|---|---|---|---|---|
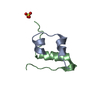
| Title | Solution structure of rat P2X4 receptor head domain | ||||||
 Components Components | P2X purinoceptor 4 | ||||||
 Keywords Keywords |  TRANSPORT PROTEIN / P2X4 receptor / TRANSPORT PROTEIN / P2X4 receptor /  head domain / CANION TRANSPORT / head domain / CANION TRANSPORT /  METAL ION BINDING METAL ION BINDING | ||||||
| Function / homology |  Function and homology information Function and homology informationPlatelet homeostasis / ligand-gated monoatomic cation channel activity / Elevation of cytosolic Ca2+ levels / sensory perception of touch / purinergic nucleotide receptor activity / extracellularly ATP-gated monoatomic cation channel activity / purinergic nucleotide receptor signaling pathway / positive regulation of microglial cell migration / negative regulation of cardiac muscle hypertrophy / ligand-gated calcium channel activity ...Platelet homeostasis / ligand-gated monoatomic cation channel activity / Elevation of cytosolic Ca2+ levels / sensory perception of touch / purinergic nucleotide receptor activity / extracellularly ATP-gated monoatomic cation channel activity / purinergic nucleotide receptor signaling pathway / positive regulation of microglial cell migration / negative regulation of cardiac muscle hypertrophy / ligand-gated calcium channel activity / response to fluid shear stress / ATP-gated ion channel activity /  regulation of chemotaxis / inorganic cation transmembrane transport / relaxation of cardiac muscle / cellular response to zinc ion / positive regulation of endothelial cell chemotaxis / cellular response to ATP / positive regulation of calcium ion transport into cytosol / response to ATP / behavioral response to pain / regulation of chemotaxis / inorganic cation transmembrane transport / relaxation of cardiac muscle / cellular response to zinc ion / positive regulation of endothelial cell chemotaxis / cellular response to ATP / positive regulation of calcium ion transport into cytosol / response to ATP / behavioral response to pain /  membrane depolarization / response to axon injury / neuronal action potential / regulation of cardiac muscle contraction / positive regulation of blood vessel endothelial cell migration / positive regulation of calcium-mediated signaling / regulation of sodium ion transport / monoatomic ion transmembrane transport / sensory perception of pain / nitric oxide biosynthetic process / membrane depolarization / response to axon injury / neuronal action potential / regulation of cardiac muscle contraction / positive regulation of blood vessel endothelial cell migration / positive regulation of calcium-mediated signaling / regulation of sodium ion transport / monoatomic ion transmembrane transport / sensory perception of pain / nitric oxide biosynthetic process /  excitatory postsynaptic potential / response to ischemia / calcium ion transmembrane transport / apoptotic signaling pathway / excitatory postsynaptic potential / response to ischemia / calcium ion transmembrane transport / apoptotic signaling pathway /  terminal bouton / terminal bouton /  regulation of blood pressure / regulation of blood pressure /  vasodilation / calcium ion transport / apical part of cell / vasodilation / calcium ion transport / apical part of cell /  cell junction / cell junction /  cell body / cell body /  dendritic spine / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / dendritic spine / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction /  cadherin binding / copper ion binding / lysosomal membrane / cadherin binding / copper ion binding / lysosomal membrane /  axon / axon /  signaling receptor binding / neuronal cell body / signaling receptor binding / neuronal cell body /  dendrite / perinuclear region of cytoplasm / dendrite / perinuclear region of cytoplasm /  signal transduction / zinc ion binding / signal transduction / zinc ion binding /  ATP binding / ATP binding /  membrane / identical protein binding / membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | ||||||
| Biological species |   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) | ||||||
| Method |  SOLUTION NMR / torsion angle dynamics SOLUTION NMR / torsion angle dynamics | ||||||
| Model details | lowest energy, model1 | ||||||
 Authors Authors | Abe, Y. / Igawa, T. / Tsuda, M. / Inoue, K. / Ueda, T. | ||||||
 Citation Citation |  Journal: FEBS Lett. / Year: 2015 Journal: FEBS Lett. / Year: 2015Title: Solution structure of the rat P2X4 receptor head domain involved in inhibitory metal binding Authors: Igawa, T. / Abe, Y. / Tsuda, M. / Inoue, K. / Ueda, T. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2rup.cif.gz 2rup.cif.gz | 358.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2rup.ent.gz pdb2rup.ent.gz | 306.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2rup.json.gz 2rup.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ru/2rup https://data.pdbj.org/pub/pdb/validation_reports/ru/2rup ftp://data.pdbj.org/pub/pdb/validation_reports/ru/2rup ftp://data.pdbj.org/pub/pdb/validation_reports/ru/2rup | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 6066.719 Da / Num. of mol.: 1 / Fragment: UNP residues 111-167 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Rattus norvegicus (Norway rat) / Gene: P2rx4 / Production host: Rattus norvegicus (Norway rat) / Gene: P2rx4 / Production host:   Escherichia coli (E. coli) / References: UniProt: P51577 Escherichia coli (E. coli) / References: UniProt: P51577 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 0.5 mM [U-99% 13C; U-99% 15N] entity-1, 50 mM Urea-2, 10 mM sodium acetate-3, 90% H2O/10% D2O Solvent system: 90% H2O/10% D2O | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||
| Sample conditions | Ionic strength: 0 / pH: 5.0 / Pressure: ambient / Temperature: 297 K |
-NMR measurement
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model : INOVA / Field strength: 600 MHz : INOVA / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
|
|---|
 Movie
Movie Controller
Controller









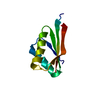


 PDBj
PDBj