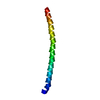
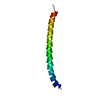
Entry Database : PDB / ID : 2rmzTitle Bicelle-embedded integrin beta3 transmembrane segment Integrin beta-3 Keywords / / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / Model type details minimized average Authors Lau, T.L. / Partridge, A.W. / Ginsberg, M.H. / Ulmer, T.S. Journal : Biochemistry / Year : 2008Title : Structure of the Integrin beta3 Transmembrane Segment in Phospholipid Bicelles and Detergent MicellesAuthors : Lau, T.L. / Partridge, A.W. / Ginsberg, M.H. / Ulmer, T.S. History Deposition Dec 4, 2007 Deposition site / Processing site Revision 1.0 Mar 18, 2008 Provider / Type Revision 1.1 Jul 13, 2011 Group Revision 1.2 Nov 10, 2021 Group / Database references / Derived calculationsCategory database_2 / pdbx_nmr_software ... database_2 / pdbx_nmr_software / pdbx_nmr_spectrometer / pdbx_struct_assembly / pdbx_struct_oper_list / struct_ref_seq_dif Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_nmr_software.name / _pdbx_nmr_spectrometer.model / _struct_ref_seq_dif.details Revision 1.3 Dec 21, 2022 Group / Category / Item Revision 1.4 May 29, 2024 Group / Category / chem_comp_bond
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Integrin beta 3
Integrin beta 3  Keywords
Keywords CELL ADHESION /
CELL ADHESION /  transmembrane helix /
transmembrane helix /  Alternative splicing / Disease mutation /
Alternative splicing / Disease mutation /  Glycoprotein / Host-virus interaction /
Glycoprotein / Host-virus interaction /  Integrin /
Integrin /  Phosphoprotein / Polymorphism / Receptor
Phosphoprotein / Polymorphism / Receptor Function and homology information
Function and homology information fibrinogen binding / glycinergic synapse / alphav-beta3 integrin-HMGB1 complex /
fibrinogen binding / glycinergic synapse / alphav-beta3 integrin-HMGB1 complex /  blood coagulation, fibrin clot formation / negative regulation of lipid transport /
blood coagulation, fibrin clot formation / negative regulation of lipid transport /  vascular endothelial growth factor receptor 2 binding / negative regulation of low-density lipoprotein receptor activity / regulation of release of sequestered calcium ion into cytosol / angiogenesis involved in wound healing / Elastic fibre formation / cell-substrate junction assembly / mesodermal cell differentiation / alphav-beta3 integrin-IGF-1-IGF1R complex /
vascular endothelial growth factor receptor 2 binding / negative regulation of low-density lipoprotein receptor activity / regulation of release of sequestered calcium ion into cytosol / angiogenesis involved in wound healing / Elastic fibre formation / cell-substrate junction assembly / mesodermal cell differentiation / alphav-beta3 integrin-IGF-1-IGF1R complex /  platelet-derived growth factor receptor binding / filopodium membrane / positive regulation of fibroblast migration /
platelet-derived growth factor receptor binding / filopodium membrane / positive regulation of fibroblast migration /  extracellular matrix binding / regulation of postsynaptic neurotransmitter receptor internalization / positive regulation of vascular endothelial growth factor receptor signaling pathway / apolipoprotein A-I-mediated signaling pathway /
extracellular matrix binding / regulation of postsynaptic neurotransmitter receptor internalization / positive regulation of vascular endothelial growth factor receptor signaling pathway / apolipoprotein A-I-mediated signaling pathway /  regulation of bone resorption / apoptotic cell clearance /
regulation of bone resorption / apoptotic cell clearance /  wound healing, spreading of epidermal cells / positive regulation of cell adhesion mediated by integrin / heterotypic cell-cell adhesion /
wound healing, spreading of epidermal cells / positive regulation of cell adhesion mediated by integrin / heterotypic cell-cell adhesion /  integrin complex / Molecules associated with elastic fibres / positive regulation of cell-matrix adhesion / cellular response to insulin-like growth factor stimulus / smooth muscle cell migration / microvillus membrane / cell adhesion mediated by integrin / Syndecan interactions / negative chemotaxis / p130Cas linkage to MAPK signaling for integrins / activation of protein kinase activity / cellular response to platelet-derived growth factor stimulus / cell-substrate adhesion /
integrin complex / Molecules associated with elastic fibres / positive regulation of cell-matrix adhesion / cellular response to insulin-like growth factor stimulus / smooth muscle cell migration / microvillus membrane / cell adhesion mediated by integrin / Syndecan interactions / negative chemotaxis / p130Cas linkage to MAPK signaling for integrins / activation of protein kinase activity / cellular response to platelet-derived growth factor stimulus / cell-substrate adhesion /  protein disulfide isomerase activity / positive regulation of smooth muscle cell migration / positive regulation of osteoblast proliferation / TGF-beta receptor signaling activates SMADs / PECAM1 interactions / lamellipodium membrane / GRB2:SOS provides linkage to MAPK signaling for Integrins / negative regulation of macrophage derived foam cell differentiation / negative regulation of lipid storage / platelet-derived growth factor receptor signaling pathway /
protein disulfide isomerase activity / positive regulation of smooth muscle cell migration / positive regulation of osteoblast proliferation / TGF-beta receptor signaling activates SMADs / PECAM1 interactions / lamellipodium membrane / GRB2:SOS provides linkage to MAPK signaling for Integrins / negative regulation of macrophage derived foam cell differentiation / negative regulation of lipid storage / platelet-derived growth factor receptor signaling pathway /  fibronectin binding / ECM proteoglycans / positive regulation of T cell migration / positive regulation of bone resorption / Integrin cell surface interactions /
fibronectin binding / ECM proteoglycans / positive regulation of T cell migration / positive regulation of bone resorption / Integrin cell surface interactions /  coreceptor activity / negative regulation of endothelial cell apoptotic process / positive regulation of substrate adhesion-dependent cell spreading /
coreceptor activity / negative regulation of endothelial cell apoptotic process / positive regulation of substrate adhesion-dependent cell spreading /  cell adhesion molecule binding / positive regulation of endothelial cell proliferation /
cell adhesion molecule binding / positive regulation of endothelial cell proliferation /  embryo implantation / positive regulation of endothelial cell migration / Integrin signaling / substrate adhesion-dependent cell spreading / cell-matrix adhesion / response to activity / Signal transduction by L1 / integrin-mediated signaling pathway / regulation of actin cytoskeleton organization /
embryo implantation / positive regulation of endothelial cell migration / Integrin signaling / substrate adhesion-dependent cell spreading / cell-matrix adhesion / response to activity / Signal transduction by L1 / integrin-mediated signaling pathway / regulation of actin cytoskeleton organization /  protein kinase C binding / positive regulation of smooth muscle cell proliferation / Signaling by high-kinase activity BRAF mutants /
protein kinase C binding / positive regulation of smooth muscle cell proliferation / Signaling by high-kinase activity BRAF mutants /  wound healing / MAP2K and MAPK activation /
wound healing / MAP2K and MAPK activation /  platelet activation /
platelet activation /  platelet aggregation / ruffle membrane / VEGFA-VEGFR2 Pathway / cellular response to mechanical stimulus / positive regulation of angiogenesis / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants /
platelet aggregation / ruffle membrane / VEGFA-VEGFR2 Pathway / cellular response to mechanical stimulus / positive regulation of angiogenesis / Signaling by RAF1 mutants / Signaling by moderate kinase activity BRAF mutants / Paradoxical activation of RAF signaling by kinase inactive BRAF / Signaling downstream of RAS mutants /  regulation of protein localization / positive regulation of peptidyl-tyrosine phosphorylation / positive regulation of fibroblast proliferation / Signaling by BRAF and RAF1 fusions
regulation of protein localization / positive regulation of peptidyl-tyrosine phosphorylation / positive regulation of fibroblast proliferation / Signaling by BRAF and RAF1 fusions
 Homo sapiens (human)
Homo sapiens (human) SOLUTION NMR /
SOLUTION NMR /  simulated annealing
simulated annealing  Authors
Authors Citation
Citation Journal: Biochemistry / Year: 2008
Journal: Biochemistry / Year: 2008 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2rmz.cif.gz
2rmz.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2rmz.ent.gz
pdb2rmz.ent.gz PDB format
PDB format 2rmz.json.gz
2rmz.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/rm/2rmz
https://data.pdbj.org/pub/pdb/validation_reports/rm/2rmz ftp://data.pdbj.org/pub/pdb/validation_reports/rm/2rmz
ftp://data.pdbj.org/pub/pdb/validation_reports/rm/2rmz Links
Links Assembly
Assembly
 Components
Components Integrin beta 3 / Platelet membrane glycoprotein IIIa / GPIIIa / CD61 antigen
Integrin beta 3 / Platelet membrane glycoprotein IIIa / GPIIIa / CD61 antigen
 Homo sapiens (human) / Gene: ITGB3, GP3A / Production host:
Homo sapiens (human) / Gene: ITGB3, GP3A / Production host: 
 Escherichia coli (E. coli) / References: UniProt: P05106
Escherichia coli (E. coli) / References: UniProt: P05106 SOLUTION NMR
SOLUTION NMR Sample preparation
Sample preparation : AVANCE / Field strength: 700 MHz
: AVANCE / Field strength: 700 MHz Processing
Processing simulated annealing / Software ordinal: 1
simulated annealing / Software ordinal: 1  Movie
Movie Controller
Controller












 PDBj
PDBj











