Entry Database : PDB / ID : 2kcvTitle Solution nmr structure of tetratricopeptide repeat domain protein sru_0103 from salinibacter ruber, northeast structural genomics consortium (nesg) target srr115c Tetratricopeptide repeat domain protein Keywords / / / / / / / / / / Function / homology Biological species Salinibacter ruber DSM 13855 (bacteria)Method / Model details lowest energy, model 1 Authors Liu, G. / Rossi, P. / Wang, D. / Nwosu, C. / Owens, L. / Xiao, R. / Liu, J. / Baran, M.C. / Swapna, G. / Acton, T.B. ...Liu, G. / Rossi, P. / Wang, D. / Nwosu, C. / Owens, L. / Xiao, R. / Liu, J. / Baran, M.C. / Swapna, G. / Acton, T.B. / Rost, B. / Montelione, G.T. / Northeast Structural Genomics Consortium (NESG) Journal : To be Published Title : Solution nmr structure of tetratricopeptide repeat domain protein sru_0103 from salinibacter ruber, northeast structural genomics consortium (nesg) target srr115cAuthors : Liu, G. / Montelione, G.T. History Deposition Dec 29, 2008 Deposition site / Processing site Revision 1.0 Jan 20, 2009 Provider / Type Revision 1.1 Jul 13, 2011 Group Revision 1.2 Mar 16, 2022 Group / Database references / Derived calculationsCategory database_2 / pdbx_nmr_software ... database_2 / pdbx_nmr_software / pdbx_nmr_spectrometer / pdbx_struct_assembly / pdbx_struct_oper_list Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_nmr_software.name / _pdbx_nmr_spectrometer.model Revision 1.3 May 22, 2024 Group / Category / chem_comp_bond
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
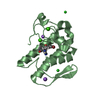
Components Tetratricopeptide repeat
Tetratricopeptide repeat  Keywords
Keywords structural genomics / unknown function /
structural genomics / unknown function /  TETRATRICOPEPTIDE REPEAT / TPR / GFT NMR / RDC / NESG / PSI-2 /
TETRATRICOPEPTIDE REPEAT / TPR / GFT NMR / RDC / NESG / PSI-2 /  Protein Structure Initiative / Northeast Structural Genomics Consortium / NUCLEOTIDE-BINDING PROTEIN
Protein Structure Initiative / Northeast Structural Genomics Consortium / NUCLEOTIDE-BINDING PROTEIN Function and homology information
Function and homology information Tetratricopeptide repeat /
Tetratricopeptide repeat /  Tetratricopeptide repeat domain /
Tetratricopeptide repeat domain /  Tetratricopeptide repeat / TPR repeat profile. /
Tetratricopeptide repeat / TPR repeat profile. /  Tetratricopeptide repeats /
Tetratricopeptide repeats /  Tetratricopeptide repeat / Serine Threonine Protein Phosphatase 5, Tetratricopeptide repeat /
Tetratricopeptide repeat / Serine Threonine Protein Phosphatase 5, Tetratricopeptide repeat /  Alpha Horseshoe / Tetratricopeptide-like helical domain superfamily / Mainly Alpha
Alpha Horseshoe / Tetratricopeptide-like helical domain superfamily / Mainly Alpha

 Salinibacter ruber DSM 13855 (bacteria)
Salinibacter ruber DSM 13855 (bacteria) SOLUTION NMR / distance geometry, torsion angle dynamics,
SOLUTION NMR / distance geometry, torsion angle dynamics,  simulated annealing,
simulated annealing,  molecular dynamics
molecular dynamics  Authors
Authors Citation
Citation Journal: To be Published
Journal: To be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2kcv.cif.gz
2kcv.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2kcv.ent.gz
pdb2kcv.ent.gz PDB format
PDB format 2kcv.json.gz
2kcv.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/kc/2kcv
https://data.pdbj.org/pub/pdb/validation_reports/kc/2kcv ftp://data.pdbj.org/pub/pdb/validation_reports/kc/2kcv
ftp://data.pdbj.org/pub/pdb/validation_reports/kc/2kcv Links
Links Assembly
Assembly
 Components
Components Tetratricopeptide repeat
Tetratricopeptide repeat

 Salinibacter ruber DSM 13855 (bacteria)
Salinibacter ruber DSM 13855 (bacteria)
 Escherichia coli (E. coli) / References: UniProt: Q2S6C5
Escherichia coli (E. coli) / References: UniProt: Q2S6C5 SOLUTION NMR
SOLUTION NMR Sample preparation
Sample preparation Processing
Processing Movie
Movie Controller
Controller












 PDBj
PDBj

