+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2jw6 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
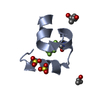
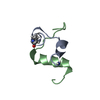
| Title | Solution structure of the DEAF1 MYND domain | |||||||||
 Components Components | Deformed epidermal autoregulatory factor 1 homolog | |||||||||
 Keywords Keywords |  TRANSCRIPTION / zinc binding domain / Disease mutation / DNA-binding / Metal-binding / TRANSCRIPTION / zinc binding domain / Disease mutation / DNA-binding / Metal-binding /  Nucleus / Nucleus /  Phosphorylation / Phosphorylation /  Secreted / Secreted /  Transcription regulation / Transcription regulation /  Zinc-finger Zinc-finger | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of mammary gland epithelial cell proliferation / embryonic skeletal system development / germ cell development / anatomical structure morphogenesis / neural tube closure / RNA polymerase II transcription regulatory region sequence-specific DNA binding /  fibrillar center / DNA-binding transcription repressor activity, RNA polymerase II-specific / transcription by RNA polymerase II / DNA-binding transcription factor activity, RNA polymerase II-specific ...regulation of mammary gland epithelial cell proliferation / embryonic skeletal system development / germ cell development / anatomical structure morphogenesis / neural tube closure / RNA polymerase II transcription regulatory region sequence-specific DNA binding / fibrillar center / DNA-binding transcription repressor activity, RNA polymerase II-specific / transcription by RNA polymerase II / DNA-binding transcription factor activity, RNA polymerase II-specific ...regulation of mammary gland epithelial cell proliferation / embryonic skeletal system development / germ cell development / anatomical structure morphogenesis / neural tube closure / RNA polymerase II transcription regulatory region sequence-specific DNA binding /  fibrillar center / DNA-binding transcription repressor activity, RNA polymerase II-specific / transcription by RNA polymerase II / DNA-binding transcription factor activity, RNA polymerase II-specific / negative regulation of DNA-templated transcription / fibrillar center / DNA-binding transcription repressor activity, RNA polymerase II-specific / transcription by RNA polymerase II / DNA-binding transcription factor activity, RNA polymerase II-specific / negative regulation of DNA-templated transcription /  chromatin / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / extracellular region / chromatin / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / extracellular region /  nucleoplasm / nucleoplasm /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing, simulated annealing,  molecular dynamics molecular dynamics | |||||||||
 Authors Authors | Spadaccini, R. / Perrin, H. / Bottomley, M. / Ansieu, S. / Sattler, M. | |||||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 2008 Journal: J.Mol.Biol. / Year: 2008Title: Retraction notice to "Structure and functional analysis of the MYND domain" [J. Mol. Biol. (2006) 358, 498-508]. Authors: Spadaccini, R. / Perrin, H. / Bottomley, M.J. / Ansieau, S. / Sattler, M. #1: Journal: J.Mol.Biol. / Year: 2006 Title: Structure and functional analysis of the MYND domain. Authors: Spadaccini, R. / Perrin, H. / Bottomley, M.J. / Ansieau, S. / Sattler, M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2jw6.cif.gz 2jw6.cif.gz | 144 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2jw6.ent.gz pdb2jw6.ent.gz | 116.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2jw6.json.gz 2jw6.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jw/2jw6 https://data.pdbj.org/pub/pdb/validation_reports/jw/2jw6 ftp://data.pdbj.org/pub/pdb/validation_reports/jw/2jw6 ftp://data.pdbj.org/pub/pdb/validation_reports/jw/2jw6 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 5878.600 Da / Num. of mol.: 1 / Fragment: mynd domain Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Plasmid: pet24d / Production host: Homo sapiens (human) / Plasmid: pet24d / Production host:   Escherichia coli (E. coli) / References: UniProt: O75398 Escherichia coli (E. coli) / References: UniProt: O75398 |
|---|---|
| #2: Chemical |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details | Contents: 0,4 mM [U-100% 13C; U-100% 15N] mynd, 4 mM DTT, 20 mM sodium phosphate, 100 mM sodium chloride, 90% H2O, 10% D2O Solvent system: 90% H2O/10% D2O | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||
| Sample conditions | Ionic strength: 20mM phosphate, 100mM NaCl / pH: 6.8 / Pressure: ambient atm / Temperature: 295 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing, simulated annealing,  molecular dynamics / Software ordinal: 1 molecular dynamics / Software ordinal: 1 Details: molecular dynamics The NMR ensemble has been refined in a shell of water molecules. refinement details can be found in the jrnl citation above and linge et al. structures were calculated ...Details: molecular dynamics The NMR ensemble has been refined in a shell of water molecules. refinement details can be found in the jrnl citation above and linge et al. structures were calculated with aria 1.2 in combination with cns. | ||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 100 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller











 PDBj
PDBj


