+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2k1v | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | R3/I5 relaxin chimera | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  HORMONE / HORMONE /  peptide hormone / relaxin-3 / insl5 / peptide hormone / relaxin-3 / insl5 /  chimera / Cleavage on pair of basic residues / chimera / Cleavage on pair of basic residues /  Secreted / Secreted /  SIGNALING PROTEIN SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRelaxin receptors / positive regulation of feeding behavior / G protein-coupled receptor binding /  hormone activity / G alpha (i) signalling events / G alpha (s) signalling events / extracellular region hormone activity / G alpha (i) signalling events / G alpha (s) signalling events / extracellular regionSimilarity search - Function | |||||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing simulated annealing | |||||||||
| Model details | Solution structure of the relaxin-3 B-chain / INSL5 A-chain chimeric peptide. | |||||||||
 Authors Authors | Rosengren, K. / Haugaard-Jonsson, L.M. | |||||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2008 Journal: J.Biol.Chem. / Year: 2008Title: Structure of the R3/I5 Chimeric Relaxin Peptide, a Selective GPCR135 and GPCR142 Agonist. Authors: Haugaard-Jonsson, L.M. / Hossain, M.A. / Daly, N.L. / Bathgate, R.A. / Wade, J.D. / Craik, D.J. / Rosengren, K.J. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2k1v.cif.gz 2k1v.cif.gz | 277 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2k1v.ent.gz pdb2k1v.ent.gz | 240.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2k1v.json.gz 2k1v.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/k1/2k1v https://data.pdbj.org/pub/pdb/validation_reports/k1/2k1v ftp://data.pdbj.org/pub/pdb/validation_reports/k1/2k1v ftp://data.pdbj.org/pub/pdb/validation_reports/k1/2k1v | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 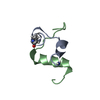
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein/peptide | Mass: 3047.586 Da / Num. of mol.: 1 / Fragment: Relaxin-3 B chain / Source method: obtained synthetically Details: Peptide chain was assembled by solid phase peptide synthesis. References: UniProt: Q8WXF3 |
|---|---|
| #2: Protein/peptide | Mass: 2202.509 Da / Num. of mol.: 1 / Fragment: Insulin-like peptide INSL5 A chain / Source method: obtained synthetically Details: Peptide chain was assembled by solid phase peptide synthesis. References: UniProt: Q9Y5Q6 |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMRDetails: Solution structure of the relaxin-3 B-chain / INSL5 A-chain chimeric peptide. | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| |||||||||||||||
| Sample conditions | Ionic strength: 0 / pH: 4 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker Avance / Manufacturer: Bruker / Model : AVANCE / Field strength: 600 MHz : AVANCE / Field strength: 600 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing / Software ordinal: 1 simulated annealing / Software ordinal: 1 Details: The structures were generated using torsion angle dynamics in CNS and subsequently refined and energy minimized in a water shell using cartesian dynamics in CNS. | ||||||||||||||||||||||||
| NMR constraints | NOE constraints total: 522 / NOE intraresidue total count: 0 / NOE long range total count: 243 / NOE medium range total count: 32 / NOE sequential total count: 247 / Hydrogen bond constraints total count: 20 | ||||||||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 50 / Conformers submitted total number: 20 / Maximum upper distance constraint violation: 0.29 Å |
 Movie
Movie Controller
Controller













 PDBj
PDBj




