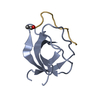
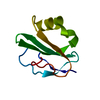
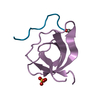
Entry Database : PDB / ID : 2jkuTitle Crystal structure of the N-terminal region of the biotin acceptor domain of human propionyl-CoA carboxylase PROPIONYL-COA CARBOXYLASE ALPHA CHAIN, MITOCHONDRIAL Keywords / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species HOMO SAPIENS (human)Method / / / Resolution : 1.5 Å Authors Healy, S. / Yue, W.W. / Kochan, G. / Pilka, E.S. / Murray, J.W. / Roos, A.K. / Filippakopoulos, P. / von Delft, F. / Arrowsmith, C. / Wikstrom, M. ...Healy, S. / Yue, W.W. / Kochan, G. / Pilka, E.S. / Murray, J.W. / Roos, A.K. / Filippakopoulos, P. / von Delft, F. / Arrowsmith, C. / Wikstrom, M. / Edwards, A. / Bountra, C. / Gravel, R.A. / Oppermann, U. Journal : Biochemistry / Year : 2010Title : Structural impact of human and Escherichia coli biotin carboxyl carrier proteins on biotin attachment.Authors : Healy, S. / McDonald, M.K. / Wu, X. / Yue, W.W. / Kochan, G. / Oppermann, U. / Gravel, R.A. History Deposition Aug 30, 2008 Deposition site / Processing site Revision 1.0 Sep 9, 2008 Provider / Type Revision 1.1 Dec 4, 2013 Group / Version format complianceRevision 1.2 Feb 28, 2018 Group / Category / citation_authorItem _citation.journal_id_ISSN / _citation.page_last ... _citation.journal_id_ISSN / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.title / _citation_author.name Revision 1.3 Mar 4, 2020 Group / OtherCategory pdbx_database_status / pdbx_struct_assembly ... pdbx_database_status / pdbx_struct_assembly / pdbx_struct_assembly_gen / pdbx_struct_assembly_prop / pdbx_struct_oper_list Item _pdbx_database_status.status_code_sf / _pdbx_struct_assembly.details ... _pdbx_database_status.status_code_sf / _pdbx_struct_assembly.details / _pdbx_struct_assembly.oligomeric_count / _pdbx_struct_assembly.oligomeric_details / _pdbx_struct_assembly_gen.oper_expression Revision 1.4 Dec 13, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_initial_refinement_model / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords LIGASE /
LIGASE /  BIOTIN / ATP-BINDING / DISEASE MUTATION / NUCLEOTIDE-BINDING /
BIOTIN / ATP-BINDING / DISEASE MUTATION / NUCLEOTIDE-BINDING /  MITOCHONDRION /
MITOCHONDRION /  PHOSPHOPROTEIN / TRANSIT PEPTIDE
PHOSPHOPROTEIN / TRANSIT PEPTIDE Function and homology information
Function and homology information propionyl-CoA carboxylase /
propionyl-CoA carboxylase /  propionyl-CoA carboxylase activity / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism /
propionyl-CoA carboxylase activity / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism /  biotin binding /
biotin binding /  catalytic complex / fatty acid metabolic process ...short-chain fatty acid catabolic process / branched-chain amino acid metabolic process / Propionyl-CoA catabolism /
catalytic complex / fatty acid metabolic process ...short-chain fatty acid catabolic process / branched-chain amino acid metabolic process / Propionyl-CoA catabolism /  propionyl-CoA carboxylase /
propionyl-CoA carboxylase /  propionyl-CoA carboxylase activity / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism /
propionyl-CoA carboxylase activity / Defective HLCS causes multiple carboxylase deficiency / Biotin transport and metabolism /  biotin binding /
biotin binding /  catalytic complex / fatty acid metabolic process /
catalytic complex / fatty acid metabolic process /  mitochondrial matrix /
mitochondrial matrix /  enzyme binding /
enzyme binding /  mitochondrion /
mitochondrion /  ATP binding /
ATP binding /  metal ion binding /
metal ion binding /  cytosol
cytosol
 HOMO SAPIENS (human)
HOMO SAPIENS (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.5 Å
MOLECULAR REPLACEMENT / Resolution: 1.5 Å  Authors
Authors Citation
Citation Journal: Biochemistry / Year: 2010
Journal: Biochemistry / Year: 2010 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2jku.cif.gz
2jku.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2jku.ent.gz
pdb2jku.ent.gz PDB format
PDB format 2jku.json.gz
2jku.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/jk/2jku
https://data.pdbj.org/pub/pdb/validation_reports/jk/2jku ftp://data.pdbj.org/pub/pdb/validation_reports/jk/2jku
ftp://data.pdbj.org/pub/pdb/validation_reports/jk/2jku Links
Links Assembly
Assembly

 Components
Components
 HOMO SAPIENS (human) / Organelle: MITOCHONDRIA
HOMO SAPIENS (human) / Organelle: MITOCHONDRIA Mitochondrion / Plasmid: PNIC28-BSA4 / Production host:
Mitochondrion / Plasmid: PNIC28-BSA4 / Production host: 
 ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / Variant (production host): R3-PRARE2 / References: UniProt: P05165,
ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / Variant (production host): R3-PRARE2 / References: UniProt: P05165,  propionyl-CoA carboxylase
propionyl-CoA carboxylase Polyethylene glycol
Polyethylene glycol Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  SLS
SLS  / Beamline: X10SA / Wavelength: 0.97628
/ Beamline: X10SA / Wavelength: 0.97628  : 0.97628 Å / Relative weight: 1
: 0.97628 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller










 PDBj
PDBj







