[English] 日本語
 Yorodumi
Yorodumi- PDB-2i0l: X-ray crystal structure of Sap97 PDZ2 bound to the C-terminal pep... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2i0l | ||||||
|---|---|---|---|---|---|---|---|
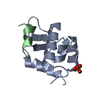
| Title | X-ray crystal structure of Sap97 PDZ2 bound to the C-terminal peptide of HPV18 E6. | ||||||
 Components Components |
| ||||||
 Keywords Keywords | PEPTIDE BINDING PROTEIN / SAP97 PDZ2 / HPV18 E6 /  tumor suppressor / tumor suppressor /  carcinoma carcinoma | ||||||
| Function / homology |  Function and homology information Function and homology informationtissue morphogenesis / regulation of protein localization to synapse / regulation of potassium ion import /  L27 domain binding / regulation of potassium ion export across plasma membrane / MPP7-DLG1-LIN7 complex / membrane raft organization / hard palate development / establishment of centrosome localization / negative regulation of p38MAPK cascade ...tissue morphogenesis / regulation of protein localization to synapse / regulation of potassium ion import / L27 domain binding / regulation of potassium ion export across plasma membrane / MPP7-DLG1-LIN7 complex / membrane raft organization / hard palate development / establishment of centrosome localization / negative regulation of p38MAPK cascade ...tissue morphogenesis / regulation of protein localization to synapse / regulation of potassium ion import /  L27 domain binding / regulation of potassium ion export across plasma membrane / MPP7-DLG1-LIN7 complex / membrane raft organization / hard palate development / establishment of centrosome localization / negative regulation of p38MAPK cascade / cortical microtubule organization / embryonic skeletal system morphogenesis / astral microtubule organization / structural constituent of postsynaptic density / lateral loop / reproductive structure development / myelin sheath abaxonal region / immunological synapse formation / L27 domain binding / regulation of potassium ion export across plasma membrane / MPP7-DLG1-LIN7 complex / membrane raft organization / hard palate development / establishment of centrosome localization / negative regulation of p38MAPK cascade / cortical microtubule organization / embryonic skeletal system morphogenesis / astral microtubule organization / structural constituent of postsynaptic density / lateral loop / reproductive structure development / myelin sheath abaxonal region / immunological synapse formation /  peristalsis / cell projection membrane / paranode region of axon / smooth muscle tissue development / bicellular tight junction assembly / Trafficking of AMPA receptors / positive regulation of potassium ion transport / Activation of Ca-permeable Kainate Receptor / peristalsis / cell projection membrane / paranode region of axon / smooth muscle tissue development / bicellular tight junction assembly / Trafficking of AMPA receptors / positive regulation of potassium ion transport / Activation of Ca-permeable Kainate Receptor /  node of Ranvier / regulation of ventricular cardiac muscle cell action potential / protein-containing complex localization / establishment or maintenance of epithelial cell apical/basal polarity / amyloid precursor protein metabolic process / endothelial cell proliferation / RAF/MAP kinase cascade / Synaptic adhesion-like molecules / neurotransmitter receptor localization to postsynaptic specialization membrane / lens development in camera-type eye / ureteric bud development / node of Ranvier / regulation of ventricular cardiac muscle cell action potential / protein-containing complex localization / establishment or maintenance of epithelial cell apical/basal polarity / amyloid precursor protein metabolic process / endothelial cell proliferation / RAF/MAP kinase cascade / Synaptic adhesion-like molecules / neurotransmitter receptor localization to postsynaptic specialization membrane / lens development in camera-type eye / ureteric bud development /  regulation of myelination / cortical actin cytoskeleton organization / branching involved in ureteric bud morphogenesis / negative regulation of G1/S transition of mitotic cell cycle / positive regulation of actin filament polymerization / receptor clustering / regulation of myelination / cortical actin cytoskeleton organization / branching involved in ureteric bud morphogenesis / negative regulation of G1/S transition of mitotic cell cycle / positive regulation of actin filament polymerization / receptor clustering /  postsynaptic density, intracellular component / postsynaptic density, intracellular component /  microvillus / microvillus /  kinesin binding / kinesin binding /  immunological synapse / immunological synapse /  basement membrane / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / lateral plasma membrane / Unblocking of NMDA receptors, glutamate binding and activation / bicellular tight junction / potassium channel regulator activity / basement membrane / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / lateral plasma membrane / Unblocking of NMDA receptors, glutamate binding and activation / bicellular tight junction / potassium channel regulator activity /  phosphatase binding / T cell proliferation / cellular response to brain-derived neurotrophic factor stimulus / negative regulation of T cell proliferation / phosphatase binding / T cell proliferation / cellular response to brain-derived neurotrophic factor stimulus / negative regulation of T cell proliferation /  T-tubule / T-tubule /  ionotropic glutamate receptor binding / actin filament polymerization / ionotropic glutamate receptor binding / actin filament polymerization /  regulation of membrane potential / regulation of membrane potential /  T cell activation / basal plasma membrane / phosphatidylinositol 3-kinase/protein kinase B signal transduction / T cell activation / basal plasma membrane / phosphatidylinositol 3-kinase/protein kinase B signal transduction /  synaptic membrane / actin filament organization / synaptic membrane / actin filament organization /  PDZ domain binding / protein localization to plasma membrane / postsynaptic density membrane / positive regulation of protein localization to plasma membrane / PDZ domain binding / protein localization to plasma membrane / postsynaptic density membrane / positive regulation of protein localization to plasma membrane /  neuromuscular junction / neuromuscular junction /  protein localization / protein localization /  : / negative regulation of ERK1 and ERK2 cascade / cytoplasmic side of plasma membrane / : / negative regulation of ERK1 and ERK2 cascade / cytoplasmic side of plasma membrane /  cell-cell adhesion / synaptic vesicle membrane / cerebral cortex development / cell-cell adhesion / synaptic vesicle membrane / cerebral cortex development /  regulation of protein localization / negative regulation of epithelial cell proliferation / cell-cell junction / regulation of protein localization / negative regulation of epithelial cell proliferation / cell-cell junction /  cell junction / cell junction /  presynaptic membrane / regulation of cell shape / chemical synaptic transmission / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / presynaptic membrane / regulation of cell shape / chemical synaptic transmission / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity /  postsynaptic membrane / symbiont-mediated perturbation of host ubiquitin-like protein modification / basolateral plasma membrane / cell population proliferation / host cell cytoplasm / postsynaptic membrane / symbiont-mediated perturbation of host ubiquitin-like protein modification / basolateral plasma membrane / cell population proliferation / host cell cytoplasm /  microtubule / transmembrane transporter binding / microtubule / transmembrane transporter binding /  postsynaptic density / molecular adaptor activity / postsynaptic density / molecular adaptor activity /  cell adhesion / neuron projection / cell adhesion / neuron projection /  membrane raft / apical plasma membrane / negative regulation of DNA-templated transcription membrane raft / apical plasma membrane / negative regulation of DNA-templated transcriptionSimilarity search - Function | ||||||
| Biological species |   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.31 Å MOLECULAR REPLACEMENT / Resolution: 2.31 Å | ||||||
 Authors Authors | Chen, X.S. / Zhang, Y. / Dasgupta, J. / Banks, L. / Thomas, M. | ||||||
 Citation Citation |  Journal: J.Virol. / Year: 2007 Journal: J.Virol. / Year: 2007Title: Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein. Authors: Zhang, Y. / Dasgupta, J. / Ma, R.Z. / Banks, L. / Thomas, M. / Chen, X.S. #1: Journal: Oncogene / Year: 2001 Title: HPV E6 and MAGUK protein interactions: determination of the molecular basis for specific protein recognition and degradation. Authors: Thomas, M. / Glaunsinger, B. / Pim, D. / Javier, R. / Banks, L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2i0l.cif.gz 2i0l.cif.gz | 49.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2i0l.ent.gz pdb2i0l.ent.gz | 35.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2i0l.json.gz 2i0l.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/i0/2i0l https://data.pdbj.org/pub/pdb/validation_reports/i0/2i0l ftp://data.pdbj.org/pub/pdb/validation_reports/i0/2i0l ftp://data.pdbj.org/pub/pdb/validation_reports/i0/2i0l | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 8843.124 Da / Num. of mol.: 2 / Fragment: PDZ2 domain Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Rattus norvegicus (Norway rat) / Production host: Rattus norvegicus (Norway rat) / Production host:   Escherichia coli (E. coli) / References: UniProt: Q62696 Escherichia coli (E. coli) / References: UniProt: Q62696#2: Protein/peptide | Mass: 947.074 Da / Num. of mol.: 2 / Source method: obtained synthetically Details: The sequence of this peptide can be found in human papillomavirus type 18 (virus) References: UniProt: P06463 #3: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.29 Å3/Da / Density % sol: 46.18 % |
|---|---|
Crystal grow | Temperature: 291 K / Method: vapor diffusion / pH: 8 Details: 2M Ammonium Sulfate, 0.1M sodium citrate, pH 8.0, VAPOR DIFFUSION, temperature 291K |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ALS ALS  / Beamline: 5.0.2 / Wavelength: 1.54 Å / Beamline: 5.0.2 / Wavelength: 1.54 Å |
| Detector | Type: MAR CCD 165 mm / Detector: CCD / Date: Jun 22, 2006 |
| Radiation | Monochromator: 1.54 / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.54 Å / Relative weight: 1 : 1.54 Å / Relative weight: 1 |
| Reflection | Resolution: 2.3→50 Å / Num. all: 7989 / Num. obs: 6978 / % possible obs: 87.3 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Biso Wilson estimate: 36.5 Å2 |
| Reflection shell | Resolution: 2.3→2.38 Å / % possible all: 93.5 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT / Resolution: 2.31→26.2 Å / Rfactor Rfree error: 0.015 / Data cutoff high absF: 1318174.96 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber MOLECULAR REPLACEMENT / Resolution: 2.31→26.2 Å / Rfactor Rfree error: 0.015 / Data cutoff high absF: 1318174.96 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 79.7247 Å2 / ksol: 0.307337 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 31.1 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.31→26.2 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.31→2.44 Å / Rfactor Rfree error: 0.053 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller










 PDBj
PDBj






