+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2a7o | ||||||
|---|---|---|---|---|---|---|---|
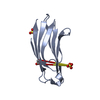
| Title | Solution Structure of the hSet2/HYPB SRI domain | ||||||
 Components Components | Huntingtin interacting protein B | ||||||
 Keywords Keywords |  TRANSCRIPTION / SRI domain / SRI / hSRI / TRANSCRIPTION / SRI domain / SRI / hSRI /  Set2 / hSet2 / phosphoCTD associating protein / Set2 Rpb1-interacting domain / PCID / PCAP Set2 / hSet2 / phosphoCTD associating protein / Set2 Rpb1-interacting domain / PCID / PCAP | ||||||
| Function / homology |  Function and homology information Function and homology informationmesoderm morphogenesis / coronary vasculature morphogenesis / morphogenesis of a branching structure / peptidyl-lysine trimethylation / cell migration involved in vasculogenesis / microtubule cytoskeleton organization involved in mitosis / [histone H3]-lysine36 N-trimethyltransferase / histone H3K36 trimethyltransferase activity / embryonic placenta morphogenesis / regulation of mRNA export from nucleus ...mesoderm morphogenesis / coronary vasculature morphogenesis / morphogenesis of a branching structure / peptidyl-lysine trimethylation / cell migration involved in vasculogenesis / microtubule cytoskeleton organization involved in mitosis / [histone H3]-lysine36 N-trimethyltransferase / histone H3K36 trimethyltransferase activity / embryonic placenta morphogenesis / regulation of mRNA export from nucleus / pericardium development / stem cell development / nucleosome organization / histone H3K36 methyltransferase activity / protein-lysine N-methyltransferase activity / response to type I interferon / embryonic cranial skeleton morphogenesis / positive regulation of ossification / response to alkaloid / regulation of protein localization to chromatin / response to metal ion / histone H3 methyltransferase activity / regulation of double-strand break repair via homologous recombination / endodermal cell differentiation / alpha-tubulin binding / positive regulation of interferon-alpha production /  mismatch repair / positive regulation of autophagy / forebrain development / mismatch repair / positive regulation of autophagy / forebrain development /  Transferases; Transferring one-carbon groups; Methyltransferases / Transferases; Transferring one-carbon groups; Methyltransferases /  regulation of cytokinesis / neural tube closure / transcription elongation by RNA polymerase II / regulation of cytokinesis / neural tube closure / transcription elongation by RNA polymerase II /  stem cell differentiation / response to organic cyclic compound / PKMTs methylate histone lysines / stem cell differentiation / response to organic cyclic compound / PKMTs methylate histone lysines /  chromosome / chromosome /  regulation of gene expression / regulation of gene expression /  angiogenesis / defense response to virus / regulation of DNA-templated transcription / angiogenesis / defense response to virus / regulation of DNA-templated transcription /  nucleoplasm / nucleoplasm /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing, simulated annealing,  molecular dynamics, torsion angle dynamics molecular dynamics, torsion angle dynamics | ||||||
 Authors Authors | Li, M. / Phatnani, H.P. / Guan, Z. / Sage, H. / Greenleaf, A. / Zhou, P. | ||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 2005 Journal: Proc.Natl.Acad.Sci.USA / Year: 2005Title: Solution structure of the Set2 Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1 Authors: Li, M. / Phatnani, H.P. / Guan, Z. / Sage, H. / Greenleaf, A. / Zhou, P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2a7o.cif.gz 2a7o.cif.gz | 650.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2a7o.ent.gz pdb2a7o.ent.gz | 546.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2a7o.json.gz 2a7o.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/a7/2a7o https://data.pdbj.org/pub/pdb/validation_reports/a7/2a7o ftp://data.pdbj.org/pub/pdb/validation_reports/a7/2a7o ftp://data.pdbj.org/pub/pdb/validation_reports/a7/2a7o | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 13147.174 Da / Num. of mol.: 1 Fragment: Set2 Rpb1-interacting (SRI) domain, HSET2/HYBP SRI domain, residues 1954-2061 Source method: isolated from a genetically manipulated source Details: isoform 1 / Source: (gene. exp.)   Homo sapiens (human) / Gene: hSet2/HYPB / Plasmid: pET-15b / Production host: Homo sapiens (human) / Gene: hSet2/HYPB / Plasmid: pET-15b / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21(DE3)STAR / References: UniProt: Q9BYW2 Escherichia coli (E. coli) / Strain (production host): BL21(DE3)STAR / References: UniProt: Q9BYW2 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions | Ionic strength: 100 mM KCl / pH: 7.0 / Pressure: ambient / Temperature: 300 K |
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiation wavelength | Relative weight: 1 | |||||||||||||||
| NMR spectrometer |
|
- Processing
Processing
| NMR software |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing, simulated annealing,  molecular dynamics, torsion angle dynamics molecular dynamics, torsion angle dynamicsSoftware ordinal: 1 Details: Structures were initially calculated using DYANA. CYANA 2.0 was used for automated data analysis to obtain additional NOE constraints. The structures were then refined against residual ...Details: Structures were initially calculated using DYANA. CYANA 2.0 was used for automated data analysis to obtain additional NOE constraints. The structures were then refined against residual dipolar couplings using XPLOR-NIH and a water-refinement protocol. | ||||||||||||||||||
| NMR representative | Selection criteria: lowest energy | ||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with favorable non-bond energy Conformers calculated total number: 25 / Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller












 PDBj
PDBj
