[English] 日本語
 Yorodumi
Yorodumi- PDB-1tut: J4/5 Loop from the Candida albicans and Candida dubliniensis Grou... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1tut | ||||||
|---|---|---|---|---|---|---|---|
| Title | J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  RNA / RNA /  internal loop / tandem mismatch / GU pairs / AA pairs internal loop / tandem mismatch / GU pairs / AA pairs | ||||||
| Function / homology |  RNA / RNA (> 10) RNA / RNA (> 10) Function and homology information Function and homology information | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  molecular dynamics molecular dynamics | ||||||
 Authors Authors | Znosko, B.M. / Kennedy, S.D. / Wille, P.C. / Krugh, T.R. / Turner, D.H. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2004 Journal: Biochemistry / Year: 2004Title: Structural Features and Thermodynamics of the J4/5 Loop from the Candida albicans and Candida dubliniensis Group I Introns. Authors: Znosko, B.M. / Kennedy, S.D. / Wille, P.C. / Krugh, T.R. / Turner, D.H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1tut.cif.gz 1tut.cif.gz | 22.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1tut.ent.gz pdb1tut.ent.gz | 14.6 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1tut.json.gz 1tut.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/tu/1tut https://data.pdbj.org/pub/pdb/validation_reports/tu/1tut ftp://data.pdbj.org/pub/pdb/validation_reports/tu/1tut ftp://data.pdbj.org/pub/pdb/validation_reports/tu/1tut | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 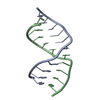
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 3648.282 Da / Num. of mol.: 1 / Source method: obtained synthetically |
|---|---|
| #2: RNA chain | Mass: 3405.034 Da / Num. of mol.: 1 / Source method: obtained synthetically |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
|
-NMR measurement
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M |
|---|---|
| Radiation wavelength | Relative weight: 1 |
| NMR spectrometer | Type: Varian INOVA / Manufacturer: Varian / Model : INOVA / Field strength: 500 MHz : INOVA / Field strength: 500 MHz |
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  molecular dynamics / Software ordinal: 1 molecular dynamics / Software ordinal: 1 Details: The structures are based on a total of 217 NMR-derived interproton distance restraints (108 intranucleotide and 109 internucleotide). Hydrogn bond restraints were used for the six WC pairs ...Details: The structures are based on a total of 217 NMR-derived interproton distance restraints (108 intranucleotide and 109 internucleotide). Hydrogn bond restraints were used for the six WC pairs and for the G8-U14 wobble pair. On the basis of NMR data that is consistent with the formation of tandem sheared AA pairs, two artificial hydrogen bonding restraints were used between each sheared AA pair. | ||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: closest to average / Conformers calculated total number: 50 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller





 PDBj
PDBj





























