+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1req | ||||||
|---|---|---|---|---|---|---|---|
| Title | METHYLMALONYL-COA MUTASE | ||||||
 Components Components | (METHYLMALONYL-COA ... ) x 2 ) x 2 | ||||||
 Keywords Keywords |  ISOMERASE / ISOMERASE /  MUTASE / INTRAMOLECULAR TRANSFERASE MUTASE / INTRAMOLECULAR TRANSFERASE | ||||||
| Function / homology |  Function and homology information Function and homology informationlactate fermentation to propionate and acetate / propionate metabolic process, methylmalonyl pathway /  methylmalonyl-CoA mutase / methylmalonyl-CoA mutase /  methylmalonyl-CoA mutase activity / methylmalonyl-CoA mutase activity /  cobalamin binding / cobalamin binding /  metal ion binding / metal ion binding /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Propionibacterium freudenreichii subsp. shermanii (bacteria) Propionibacterium freudenreichii subsp. shermanii (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2 Å SYNCHROTRON / Resolution: 2 Å | ||||||
 Authors Authors | Evans, P.R. / Mancia, F. | ||||||
 Citation Citation |  Journal: Structure / Year: 1996 Journal: Structure / Year: 1996Title: How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 A resolution. Authors: Mancia, F. / Keep, N.H. / Nakagawa, A. / Leadlay, P.F. / McSweeney, S. / Rasmussen, B. / Bosecke, P. / Diat, O. / Evans, P.R. #1:  Journal: Biochem.J. / Year: 1990 Journal: Biochem.J. / Year: 1990Title: Adenosylcobalamin-Dependent Methylmalonyl-Coa Mutase from Propionibacterium Shermanii. Active Holoenzyme Produced from Escherichia Coli Authors: Mckie, N. / Keep, N.H. / Patchett, M.L. / Leadlay, P.F. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1req.cif.gz 1req.cif.gz | 573.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1req.ent.gz pdb1req.ent.gz | 453.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1req.json.gz 1req.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/re/1req https://data.pdbj.org/pub/pdb/validation_reports/re/1req ftp://data.pdbj.org/pub/pdb/validation_reports/re/1req ftp://data.pdbj.org/pub/pdb/validation_reports/re/1req | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| Unit cell |
| ||||||||
| Noncrystallographic symmetry (NCS) | NCS oper: (Code: given Matrix: (-0.464135, 0.013887, 0.885656), Vector  : : Details | THE ASYMMETRIC UNIT OF THE CRYSTAL CONTAINS TWO HETERODIMERIC MOLECULES, EACH WITH AN ALPHA CHAIN (CHAINS A AND C, CORRESPONDING TO GENE MUTB) AND A BETA CHAIN (CHAINS B AND D, CORRESPONDING TO GENE MUTA). MOLECULE 1 CONSISTS OF CHAINS A (ALPHA), B (BETA), WITH GLYCEROL CHAIN G AND WATERS X. MOLECULE 2 CONSISTS OF CHAINS C (ALPHA), D (BETA), WITH GLYCEROL CHAIN H AND WATERS Y. CHAINS A AND C INCLUDE COENZYME B12 (RESIDUE 800), DESULPHO-COA (RESIDUE 801) AND A GLYCEROL IN THE ACTIVE SITE (RESIDUE 802). | |
- Components
Components
-METHYLMALONYL-COA ... , 2 types, 4 molecules ACBD
| #1: Protein |  Mass: 80137.852 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: CHAINS A AND C INCLUDE COENZYME B12, DESULPHO-COA, AND A GLYCEROL IN THE ACTIVE SITE. B12 IS PRESENT LARGELY AS REDUCED COB(II)ALAMIN, OR B12R. Source: (gene. exp.)   Propionibacterium freudenreichii subsp. shermanii (bacteria) Propionibacterium freudenreichii subsp. shermanii (bacteria)Species: Propionibacterium freudenreichii  / Strain: NCIB 9885 / Strain: NCIB 9885Description: THE 2 GENES MUTA (BETA CHAIN) AND MUTB (ALPHA CHAIN) ARE COEXPRESSED FROM THE SAME PLASMID Cell line: 293 / Gene: MUTA MUTB / Plasmid: PMEX1 / Gene (production host): MUTA, MUTB / Production host: K38 PGP1-2 / Strain (production host): 293 / References: UniProt: P11653,  methylmalonyl-CoA mutase methylmalonyl-CoA mutase#2: Protein |  Mass: 69430.188 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Details: CHAINS A AND C INCLUDE COENZYME B12, DESULPHO-COA, AND A GLYCEROL IN THE ACTIVE SITE. B12 IS PRESENT LARGELY AS REDUCED COB(II)ALAMIN, OR B12R. Source: (gene. exp.)   Propionibacterium freudenreichii subsp. shermanii (bacteria) Propionibacterium freudenreichii subsp. shermanii (bacteria)Species: Propionibacterium freudenreichii  / Strain: NCIB 9885 / Strain: NCIB 9885Description: THE 2 GENES MUTA (BETA CHAIN) AND MUTB (ALPHA CHAIN) ARE COEXPRESSED FROM THE SAME PLASMID Cell line: 293 / Gene: MUTA MUTB / Plasmid: PMEX1 / Gene (production host): MUTA, MUTB / Production host:   Escherichia coli (E. coli) / Strain (production host): 293 / References: UniProt: P11652, Escherichia coli (E. coli) / Strain (production host): 293 / References: UniProt: P11652,  methylmalonyl-CoA mutase methylmalonyl-CoA mutase |
|---|
-Non-polymers , 4 types, 1545 molecules 
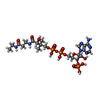





| #3: Chemical |  Vitamin B12 Vitamin B12#4: Chemical | #5: Chemical | ChemComp-GOL /  Glycerol Glycerol#6: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.76 Å3/Da / Density % sol: 48 % | ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | Details: THE CRYSTALS WERE GROWN IN THE PRESENCE OF 2MM EXCESS 5'-DEOXYADENOSYLCOBALAMIN (COENZYME B12) AND 12MM DESULPHO-COA (FINAL CONCENTRATIONS), EQUILIBRATED AGAINST 14% W/V PEG 4000 AND 20% V/V ...Details: THE CRYSTALS WERE GROWN IN THE PRESENCE OF 2MM EXCESS 5'-DEOXYADENOSYLCOBALAMIN (COENZYME B12) AND 12MM DESULPHO-COA (FINAL CONCENTRATIONS), EQUILIBRATED AGAINST 14% W/V PEG 4000 AND 20% V/V GLYCEROL, 100MM TRIS PH 7.5. THE ELECTRON DENSITY MAPS SHOW NO ADENOSYL GROUP ATTACHED TO THE COBALT ATOM, AND SPECTRA FROM SIMILAR CRYSTALS SHOW EVIDENCE OF SUBSTANTIAL REDUCTION OF COIII TO COII, WHICH IS 5-COORDINATE. THE COBALAMIN IN THIS CRYSTAL STRUCTURE IS BEST CONSIDERED AS REDUCED COB(II)ALAMIN (OR B12R). THE BOND LENGTH FROM THE COBALT TO THE LOWER AXIAL LIGAND, NE2 OF HIS A 610 (OR HIS C 610) IS SIGNIFICANTLY LONGER THAN THAT IN MODEL COMPOUNDS. POORLY ORDERED LOOPS: DENSITY IN THE FOLLOWING REGIONS IS POOR, AND THE MODEL MUST BE CONSIDERED UNRELIABLE. ALPHA CHAIN: A 1, C 1 MISSING, A 2 - A 3, C 2 - C 3 WEAK. BETA CHAIN: B 1 - C 19, C 1 - C 16 MISSING; B 184 - B 191, D 184 - D 191 WEAK; D 228 - D 230 WEAK; D 271 - D 276 VERY POOR DENSITY (MUCH BETTER IN CHAIN B); D 315 WEAK; D 474 - D 428 WEAK. | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 23 ℃ / pH: 7.5 / Method: vapor diffusion | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID2 / Wavelength: 0.9 / Beamline: ID2 / Wavelength: 0.9 |
|---|---|
| Detector | Type: MAR scanner 300 mm plate / Detector: IMAGE PLATE / Date: Jun 24, 1994 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.9 Å / Relative weight: 1 : 0.9 Å / Relative weight: 1 |
| Reflection | Resolution: 2→20 Å / Num. obs: 217377 / % possible obs: 99.8 % / Observed criterion σ(I): 5 / Redundancy: 4.6 % / Rmerge(I) obs: 0.051 |
| Reflection | *PLUS Num. measured all: 1008268 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2→20 Å / σ(F): 0 Details: DERIVED FROM ENGH AND HUBER PARAMETERS BY V. LAMZIN FINAL RMS COORD. SHIFT 0.009 ANGSTROMS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 42 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→20 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.22 / Rfactor Rwork : 0.22 : 0.22 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller













 PDBj
PDBj


